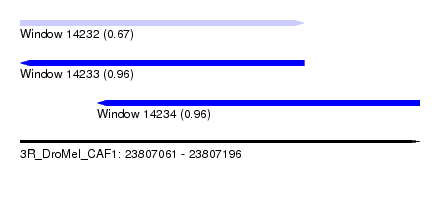
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,807,061 – 23,807,196 |
| Length | 135 |
| Max. P | 0.962916 |

| Location | 23,807,061 – 23,807,157 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.93 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23807061 96 + 27905053 UGUGUUAGGUGUGUGAG----CGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACAACCACCUGAAUUGGCGUUGGAGCAUCUGAGCCAUCAAAGAAAUUA .(..(((.(((((....----))))).)))..)...(((((...(((((((..((((..((......)).))))..))))))).)))))........... ( -32.50) >DroSec_CAF1 12354 94 + 1 UCUGUUUAG-GAGUGAG----CGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACAAC-UCCUGAAUUGGCGAUGGAGCAUCUGAGCCAUCAAAGAAAUUA (((((((((-((((.((----(((.....)))))...(((((......)))))..))-))))))))....(((((..(....)..)))))..)))..... ( -33.40) >DroSim_CAF1 12397 95 + 1 UGUGUUAAG-GAGUGAG----CGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACAACCACCUGAAUUGGCGAUGGAGCAUCUGAGCCAUCAAAGAAAUUA .........-..((.((----..((.((((((((....)))..((((...)))).))))).))..)).))(((((..(....)..))))).......... ( -25.90) >DroEre_CAF1 12601 91 + 1 ---GUUAA--GCGUGAG----CGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACCACCACCUGAAUUGGCGGUGGAGCAUCUGAGCCAUCAAAGAAAUUA ---.....--((....)----)((((...))))...(((((...(((((((..(((((.((......)).))))).))))))).)))))........... ( -34.50) >DroYak_CAF1 12904 95 + 1 ---GUUAG--GCGUGAGCGGGCGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACAACCACCUGAAUUGGCGGUGGAGCAUCUGAGCCACCAAAGAAAUUA ---.....--((....))((((((.....))))....((((...(((((((..((.((.((......)).))))..))))))).)))))).......... ( -33.90) >DroPer_CAF1 12431 89 + 1 ---GUGCG--GCAUCA------CCAUGUGGCAUCAAACGGCAAGUACCUGCCCCCUCCACCCGUUGGGGGGCAGGGCCAUCUGAGGCAAAAGCGAAACCA ---((((.--((((..------..)))).))))......((.....((((((((((.........))))))))))(((......)))....))....... ( -36.20) >consensus ___GUUAA__GCGUGAG____CGCAUGUGGUGCUAAAUGGCAAGUGGAUGCCACAACCACCUGAAUUGGCGAUGGAGCAUCUGAGCCAUCAAAGAAAUUA ......................((....((((.....(((((......)))))....)))).......))(((((..(....)..))))).......... (-14.74 = -15.93 + 1.20)

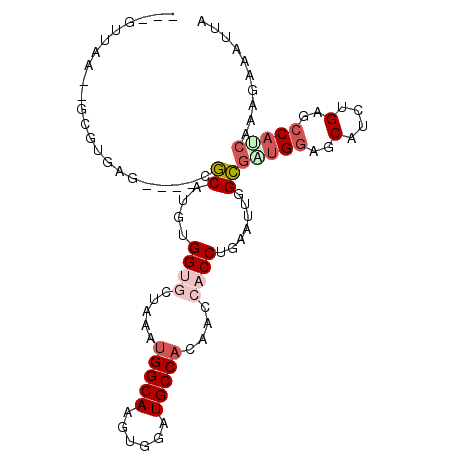
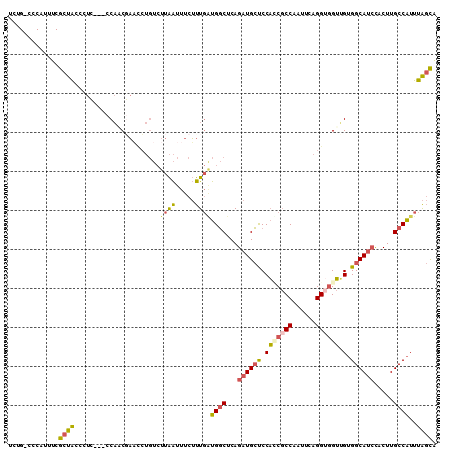
| Location | 23,807,061 – 23,807,157 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23807061 96 - 27905053 UAAUUUCUUUGAUGGCUCAGAUGCUCCAACGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCACCACAUGCG----CUCACACACCUAACACA ..........((((((...(((((..(((((((......))).))))..))))).....))))))..(((.....))).----................. ( -25.60) >DroSec_CAF1 12354 94 - 1 UAAUUUCUUUGAUGGCUCAGAUGCUCCAUCGCCAAUUCAGGA-GUUGUGGCAUCCACUUGCCAUUUAGCACCACAUGCG----CUCACUC-CUAAACAGA .......((((.((((...((((...))))))))....((((-((.((((((......))))))...(((.....))).----...))))-))...)))) ( -24.30) >DroSim_CAF1 12397 95 - 1 UAAUUUCUUUGAUGGCUCAGAUGCUCCAUCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCACCACAUGCG----CUCACUC-CUUAACACA ..........((((((...(((((..(((((((......))))).))..))))).....))))))..(((.....))).----.......-......... ( -25.40) >DroEre_CAF1 12601 91 - 1 UAAUUUCUUUGAUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUGGUGGCAUCCACUUGCCAUUUAGCACCACAUGCG----CUCACGC--UUAAC--- ..........((((((...((((((((((((((......)))))))).)))))).....))))))..(((.....)))(----(....))--.....--- ( -34.00) >DroYak_CAF1 12904 95 - 1 UAAUUUCUUUGGUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCACCACAUGCGCCCGCUCACGC--CUAAC--- ..........((((((...(((((..(((((((......))))).))..))))).....))).....(((.....)))..........))--)....--- ( -28.30) >DroPer_CAF1 12431 89 - 1 UGGUUUCGCUUUUGCCUCAGAUGGCCCUGCCCCCCAACGGGUGGAGGGGGCAGGUACUUGCCGUUUGAUGCCACAUGG------UGAUGC--CGCAC--- .(((.(((((...((.(((((((((((((((((((........).))))))))).....))))))))).)).....))------))).))--)....--- ( -44.50) >consensus UAAUUUCUUUGAUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCACCACAUGCG____CUCACGC__CUAAC___ ..........((((((...((((((.(((((((......)))))).).)))))).....))))))..(((.....)))...................... (-17.77 = -18.72 + 0.95)

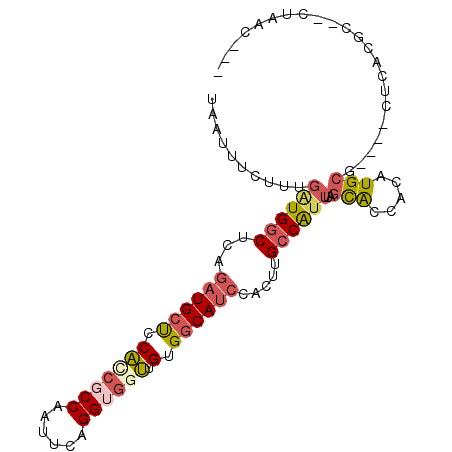
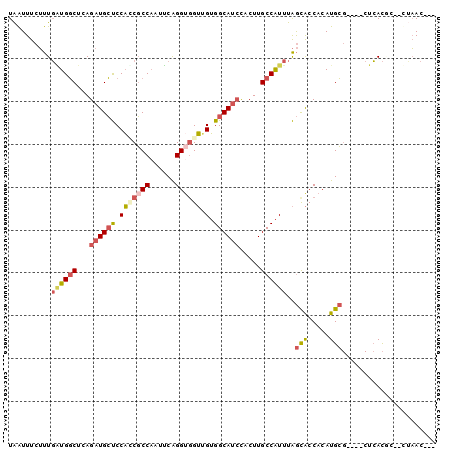
| Location | 23,807,087 – 23,807,196 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
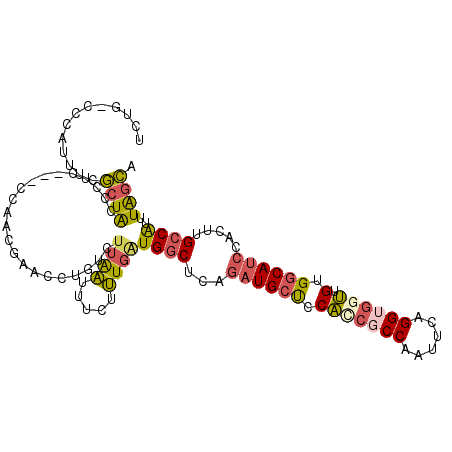
>3R_DroMel_CAF1 23807087 109 - 27905053 UCUG-CCCAUCUCGCUAACCUCGAACCAACGAUCCUGUCUUAAUUUCUUUGAUGGCUCAGAUGCUCCAACGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCA ....-........((((...(((......)))..................((((((...(((((..(((((((......))).))))..))))).....)))))))))). ( -29.10) >DroPse_CAF1 12361 102 - 1 CCCGCUUCACUUCACUGCCUUG---CCCUCGUUUC-----UGGUUUCGCUUUUGCCUCAGAUGGCCCUGCCCCCCAACGGGUGGAGGGGGCAGGUACUUGCCGUUUGAUG ................((...(---((........-----.)))...)).......(((((((((((((((((((........).))))))))).....))))))))).. ( -34.30) >DroSec_CAF1 12379 105 - 1 UCUG-CCCAUUUCGCUACCCUC---CCAACGAACCUGUCUUAAUUUCUUUGAUGGCUCAGAUGCUCCAUCGCCAAUUCAGGA-GUUGUGGCAUCCACUUGCCAUUUAGCA ....-........((((.....---.((((...((((..((((.....))))((((...((((...))))))))...)))).-))))(((((......)))))..)))). ( -24.40) >DroSim_CAF1 12422 105 - 1 UCUG-CCCAAUUCGCUAC-CUC---CCAACGAACCUGUCUUAAUUUCUUUGAUGGCUCAGAUGCUCCAUCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCA ....-........((((.-...---.(((.(((...........))).)))(((((...(((((..(((((((......))))).))..))))).....))))).)))). ( -27.00) >DroEre_CAF1 12622 106 - 1 UCUG-CCCAUUUCGCUGCCUUG---CCAUCUAACCUGUCUUAAUUUCUUUGAUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUGGUGGCAUCCACUUGCCAUUUAGCA ....-........((((....(---(((((....................))))))...((((((((((((((......)))))))).))))))...........)))). ( -36.35) >DroYak_CAF1 12929 106 - 1 ACUG-CCCAUUUCGCUACCUUC---CCAUCUAAAAUGUCUUAAUUUCUUUGGUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCA ....-........((((.....---(((....((((......))))...)))((((...(((((..(((((((......))))).))..))))).....))))..)))). ( -30.40) >consensus UCUG_CCCAUUUCGCUACCCUC___CCAACGAACCUGUCUUAAUUUCUUUGAUGGCUCAGAUGCUCCACCGCCAAUUCAGGUGGUUGUGGCAUCCACUUGCCAUUUAGCA .............((((......................((((.....))))((((...((((((.(((((((......)))))).).)))))).....))))..)))). (-18.33 = -18.50 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:38 2006