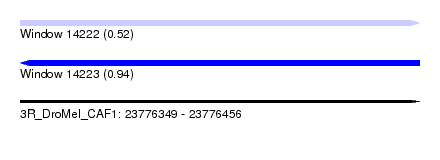
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,776,349 – 23,776,456 |
| Length | 107 |
| Max. P | 0.939397 |

| Location | 23,776,349 – 23,776,456 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.81 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.41 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23776349 107 + 27905053 CUUUGCCUGUUCAAAUAAUUAGUUAUUUAU----------GAUAAGCAUUUCCGCAGAAUCCAUUAUAAUAUGGAGAAUAGAAUGACGUGUAUUAUUUUUU--GCCAACAAAUUAAUAU .((((...((..(((((((..(((((((..----------.....((......))....(((((......))))).....)))))))....)))))))...--))...))))....... ( -15.90) >DroSec_CAF1 10844 101 + 1 --------------AUGAUUAGUUAUUUAUGGAUACUGCUGAUAAGCAUAUCCGCAGAAUCCAUUAUAAUAUUGAGAAUAG--UGACGUGUAUUAUUAUUU--GCCAGCUAUGCAAUAU --------------.......(((((..((((((.((((.((((....)))).)))).)))))).)))))((((...((((--(...(.(((........)--))).))))).)))).. ( -24.60) >DroSim_CAF1 8048 101 + 1 --------------AUGAUUAGUUAUUUAUGGAUACUGCUGAUAAGCAUUUCCGCAGAAUCCAUUAUAAUAUUGAGAAUAG--UGACGUGUAUUAUUAUUU--GCCAGCUAUGCAAUAU --------------.......(((((..((((((.((((.((........)).)))).)))))).)))))((((...((((--(...(.(((........)--))).))))).)))).. ( -23.00) >DroEre_CAF1 8070 114 + 1 UUUU-CAUACUUAAACUGGGAGUUAUUUAUGGAUCCUGCUGAUAAGAAUUUCCGCAGAAUCCAUUUUAAUAUUGACCACGG--UGACGUGUAUUCUUUUUU--GCCAGCCUUUUAUUAU ....-..........((((.(((((...((((((.((((.((........)).)))).))))))..))))......((((.--...))))..........)--.))))........... ( -28.50) >DroYak_CAF1 10915 118 + 1 AUUU-CAUGCUAAACCGGGGAAUUAUUUAUGGAUCAUGCUGAUAAGAAGUUCCGCAGAAUCCAUUUUAAUAUUGACCAUAGUGUGACGUGUAUUCUUUUUUGUGCCAGCCUUUUAUUUU ...(-(((((((.....((..(.((((.((((((..(((.((........)).)))..))))))...)))).)..)).)))))))).(.((((........)))))............. ( -25.20) >consensus _UUU_C_U__U_AAAUGAUUAGUUAUUUAUGGAUACUGCUGAUAAGCAUUUCCGCAGAAUCCAUUAUAAUAUUGAGAAUAG__UGACGUGUAUUAUUUUUU__GCCAGCCAUUUAAUAU ..................((...((((.((((((.((((.((........)).)))).))))))...))))....)).......................................... ( -9.52 = -10.36 + 0.84)

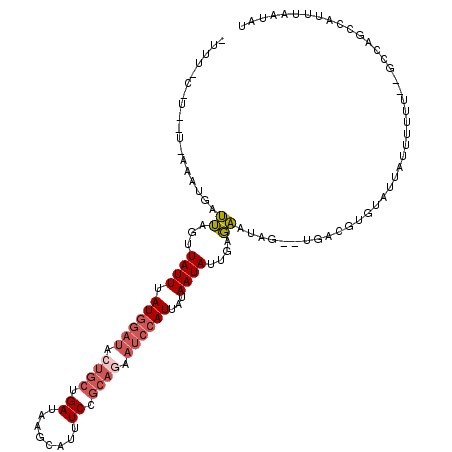
| Location | 23,776,349 – 23,776,456 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.81 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -7.80 |
| Energy contribution | -9.60 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23776349 107 - 27905053 AUAUUAAUUUGUUGGC--AAAAAAUAAUACACGUCAUUCUAUUCUCCAUAUUAUAAUGGAUUCUGCGGAAAUGCUUAUC----------AUAAAUAACUAAUUAUUUGAACAGGCAAAG .......((((((...--...(((((((................(((((......)))))....(((....))).....----------...........))))))).....)))))). ( -12.80) >DroSec_CAF1 10844 101 - 1 AUAUUGCAUAGCUGGC--AAAUAAUAAUACACGUCA--CUAUUCUCAAUAUUAUAAUGGAUUCUGCGGAUAUGCUUAUCAGCAGUAUCCAUAAAUAACUAAUCAU-------------- ((((((.((((.((((--..............))))--))))...))))))....((((((.((((.((((....)))).)))).))))))..............-------------- ( -23.24) >DroSim_CAF1 8048 101 - 1 AUAUUGCAUAGCUGGC--AAAUAAUAAUACACGUCA--CUAUUCUCAAUAUUAUAAUGGAUUCUGCGGAAAUGCUUAUCAGCAGUAUCCAUAAAUAACUAAUCAU-------------- ((((((.((((.((((--..............))))--))))...))))))....((((((.((((.((........)).)))).))))))..............-------------- ( -20.04) >DroEre_CAF1 8070 114 - 1 AUAAUAAAAGGCUGGC--AAAAAAGAAUACACGUCA--CCGUGGUCAAUAUUAAAAUGGAUUCUGCGGAAAUUCUUAUCAGCAGGAUCCAUAAAUAACUCCCAGUUUAAGUAUG-AAAA ........(((((((.--......((...((((...--.)))).)).........(((((((((((.((........)).))))))))))).........))))))).......-.... ( -30.50) >DroYak_CAF1 10915 118 - 1 AAAAUAAAAGGCUGGCACAAAAAAGAAUACACGUCACACUAUGGUCAAUAUUAAAAUGGAUUCUGCGGAACUUCUUAUCAGCAUGAUCCAUAAAUAAUUCCCCGGUUUAGCAUG-AAAU ........(((((((................(((......)))............(((((((.(((.((........)).))).)))))))..........)))))))......-.... ( -18.50) >consensus AUAUUAAAUAGCUGGC__AAAAAAUAAUACACGUCA__CUAUUCUCAAUAUUAUAAUGGAUUCUGCGGAAAUGCUUAUCAGCAGGAUCCAUAAAUAACUAAUCAUUU_A__A_G_AAA_ .......................................................((((((.((((.((........)).)))).))))))............................ ( -7.80 = -9.60 + 1.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:27 2006