
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,773,781 – 23,773,872 |
| Length | 91 |
| Max. P | 0.989427 |

| Location | 23,773,781 – 23,773,872 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -2.72 |
| Energy contribution | -2.32 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23773781 91 + 27905053 CAGCACAAGUC---------CACCCACAUGUG----GGUGCGGAACCUUAACGCCAUGGCAAAU----------------AAAAAUACCAACAUUAAAAUAUUUAACUUAACAUUGUAAA ....((((.((---------((((((....))----)))).)).........((....))....----------------.................................))))... ( -14.80) >DroSec_CAF1 8291 91 + 1 CAGCCCAAGUC---------CACCCACAUGUG----GAUGCUGAACCUUAACGCCAUGGCAAAU----------------AAAAAUACCAAUAUUAAAAUAUUUAACUUAACUUUGUAAA ((((....(((---------(((......)))----))))))).........((....))....----------------........................................ ( -14.10) >DroSim_CAF1 5466 90 + 1 CAGUCAAAGUC---------CACCCACAUGUG----GGUGC-GAACCUUAACGCCGUGGCAAAU----------------UAAAAUACCACCAUUAAAAUAUUUAACUUAACUUUGUAAA ....(((((((---------((((((....))----)))).-))...........((((.....----------------.......))))....................))))).... ( -17.60) >DroEre_CAF1 5519 120 + 1 CAAGCCAAGUCCAGAAAUGCCACCCACAUGUGGGUGGGUGCAGAACUUAAAUAUCGUGAAGAAAAAUACUUAAAUACUAGAAAAAUACCAACAUUAAAAUGUUUUACUAAACUUUAUAAA ...(((.....((....))(((((((....)))))))).))..............((((((.......((........)).........(((((....)))))........))))))... ( -19.90) >DroYak_CAF1 8319 112 + 1 CAGGCCACGUCCAGAUAUGCCACCCACAUGUG----GGUGCAGAACUUUAAUUUUGCGGAGUAUUAAAC----AUACUAGAAACGUACCAACAUUAAAAUAUUUAACUUAACAUUAUAAA ..((..((((.........(((((((....))----)))((((((......))))))))(((((.....----)))))....)))).))............................... ( -20.60) >consensus CAGCCCAAGUC_________CACCCACAUGUG____GGUGCAGAACCUUAACGCCGUGGCAAAU________________AAAAAUACCAACAUUAAAAUAUUUAACUUAACUUUGUAAA ..........................((((((....(((.....)))....)).)))).............................................................. ( -2.72 = -2.32 + -0.40)


| Location | 23,773,781 – 23,773,872 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.63 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.46 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
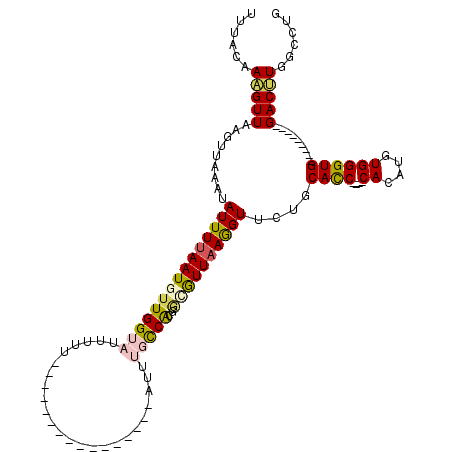
>3R_DroMel_CAF1 23773781 91 - 27905053 UUUACAAUGUUAAGUUAAAUAUUUUAAUGUUGGUAUUUUU----------------AUUUGCCAUGGCGUUAAGGUUCCGCACC----CACAUGUGGGUG---------GACUUGUGCUG ..(((((.(((.........(((((((((((((((.....----------------...)))))..))))))))))....((((----((....))))))---------))))))))... ( -25.90) >DroSec_CAF1 8291 91 - 1 UUUACAAAGUUAAGUUAAAUAUUUUAAUAUUGGUAUUUUU----------------AUUUGCCAUGGCGUUAAGGUUCAGCAUC----CACAUGUGGGUG---------GACUUGGGCUG .......(((((((((....((((((((..(((((.....----------------...)))))....))))))))....((((----((....))))))---------))))).)))). ( -20.60) >DroSim_CAF1 5466 90 - 1 UUUACAAAGUUAAGUUAAAUAUUUUAAUGGUGGUAUUUUA----------------AUUUGCCACGGCGUUAAGGUUC-GCACC----CACAUGUGGGUG---------GACUUUGACUG ....(((((((.........(((((((((((((((.....----------------...))))))..)))))))))..-.((((----((....))))))---------))))))).... ( -29.50) >DroEre_CAF1 5519 120 - 1 UUUAUAAAGUUUAGUAAAACAUUUUAAUGUUGGUAUUUUUCUAGUAUUUAAGUAUUUUUCUUCACGAUAUUUAAGUUCUGCACCCACCCACAUGUGGGUGGCAUUUCUGGACUUGGCUUG ......((((((((...(((((....)))))..........(((.(((((((((((.........))))))))))).)))...(((((((....))))))).....))))))))...... ( -27.30) >DroYak_CAF1 8319 112 - 1 UUUAUAAUGUUAAGUUAAAUAUUUUAAUGUUGGUACGUUUCUAGUAU----GUUUAAUACUCCGCAAAAUUAAAGUUCUGCACC----CACAUGUGGGUGGCAUAUCUGGACGUGGCCUG .....(((((((((........)))))))))((((((..((((((((----(((.........(((.(((....))).)))(((----((....))))))))))).)))))))).))).. ( -27.60) >consensus UUUACAAAGUUAAGUUAAAUAUUUUAAUGUUGGUAUUUUU________________AUUUGCCACGGCGUUAAGGUUCUGCACC____CACAUGUGGGUG_________GACUUGGCCUG ......(((((.........(((((((((((((((........................)))))..))))))))))....((((....((....)))))).........)))))...... ( -8.10 = -8.46 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:25 2006