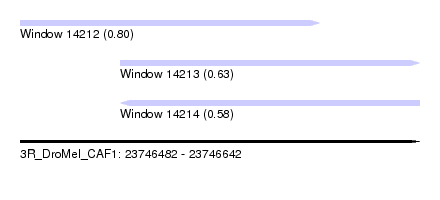
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,746,482 – 23,746,642 |
| Length | 160 |
| Max. P | 0.798432 |

| Location | 23,746,482 – 23,746,602 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -37.44 |
| Energy contribution | -37.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23746482 120 + 27905053 AUCCUCCUGCUUGGCCUUCAUCUCGUCGAGGGUGACCAGACCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAG .(((((.......((((((........))))))(((((......))))))))))((..(((((..((.((((.........))))))(((((..(((....)))..))))))))))..)) ( -43.30) >DroSec_CAF1 196203 120 + 1 AUCCUCCUGCUUGGCCUUCAUCUCGUCGAGGGUGACCAGACCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAAUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAG .(((((.......((((((........))))))(((((......))))))))))((..(((((..((((.(((....)))...))))(((((..(((....)))..))))))))))..)) ( -40.80) >DroSim_CAF1 196182 120 + 1 AUCCUCCUGCUUGGCCUUCAUCUCGUCGAGGGUGACCAGACCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAG .(((((.......((((((........))))))(((((......))))))))))((..(((((..((.((((.........))))))(((((..(((....)))..))))))))))..)) ( -43.30) >DroEre_CAF1 206076 120 + 1 AUCCUGCUGCUUGGCCUUCAUCUCGUCCAGGGUGACCAGGCCAAUGGUCGAGGACUUAAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCGAUCUUUUCCAGCUUCAG .....((((.(((((((((((((......))))))..))))))).((((((((((......(((..(.((((.........)))))..))).....)))))))))).....))))..... ( -48.80) >DroYak_CAF1 203429 120 + 1 GUCCUCCUGCUUGGCCUUCAUCUCGUCCAGGGUGACCAGACCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCGAACUUGUUCUCAAUCUUGUCCAGCUUCAG ((((((((((...(((........((((.((....)).((((...))))..))))..((((....))))...)))...)))).))).)))(((.(((..(........)..))).))).. ( -40.90) >consensus AUCCUCCUGCUUGGCCUUCAUCUCGUCGAGGGUGACCAGACCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAG ...(((..(((.(((((((........))))))(((((......)))))((((((..(((.(((..(.((((.((...)).)))))..)))..)))))))))........).)))..))) (-37.44 = -37.68 + 0.24)



| Location | 23,746,522 – 23,746,642 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -32.52 |
| Energy contribution | -32.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23746522 120 + 27905053 CCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAGCUCCUCCUCGAUCUUCUUCUUGCGAAACUCGAUCUCCUGC .....(((((((((((..(((((..((.((((.........))))))(((((..(((....)))..))))))))))..))....))))))))).........((.....))......... ( -36.20) >DroSec_CAF1 196243 120 + 1 CCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAAUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAGCUCCUCCUCGAUCUUCUUCUUGCGAAACUCGAUCUCCUGC .....(((((((((((..(((((..((((.(((....)))...))))(((((..(((....)))..))))))))))..))....))))))))).........((.....))......... ( -33.70) >DroSim_CAF1 196222 120 + 1 CCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAGCUCCUCCUCGAUCUUCUUCUUGCGGAACUCGAUCUCCUGC .....(((((((((((..(((((..((.((((.........))))))(((((..(((....)))..))))))))))..))....)))))))))........(((((........))).)) ( -38.60) >DroEre_CAF1 206116 120 + 1 CCAAUGGUCGAGGACUUAAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCGAUCUUUUCCAGCUUCAGCUCCUCCCCGAUCUUCUUCUUGCGGAAUUCGAUCUCCUGC .....((((((((((......(((..(.((((.........)))))..))).....))))))))))......(((....))).......((((...(((.....)))...))))...... ( -33.80) >DroYak_CAF1 203469 120 + 1 CCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCGAACUUGUUCUCAAUCUUGUCCAGCUUCAGCUCCUCCUCGAUCUUCUUUUUGCGGAACUCGAUCUCCUGC .....(((((((((...((((((..(((((((.........))))(.(((((..(((....)))..)))))))))..)))))).)))))))))........(((((........))).)) ( -40.20) >consensus CCAAUGGUCGAGGACUUGAGCUGCUGCUCCACGGCAUCGUAGUGGGUGGCAAACUUGUUCUCAAUCUUGUCCAGCUUGAGCUCCUCCUCGAUCUUCUUCUUGCGGAACUCGAUCUCCUGC .....(((((((((((((((((((..(.((((.((...)).)))))..))....(((....)))........)))))))).)))))...((..(((.......)))..))))))...... (-32.52 = -32.56 + 0.04)


| Location | 23,746,522 – 23,746,642 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -33.04 |
| Energy contribution | -32.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23746522 120 - 27905053 GCAGGAGAUCGAGUUUCGCAAGAAGAAGAUCGAGGAGGAGCUCAAGCUGGACAAGAUUGAGAACAAGUUUGCCACCCACUACGAUGCCGUGGAGCAGCAGCUCAAGUCCUCGACCAUUGG ...((.((((...((((....))))..))))(((((.(((((...(((((.((((.(((....))).)))))).....(((((....))))))))...)))))...)))))..))..... ( -41.60) >DroSec_CAF1 196243 120 - 1 GCAGGAGAUCGAGUUUCGCAAGAAGAAGAUCGAGGAGGAGCUCAAGCUGGACAAGAUUGAGAACAAGUUUGCCACCCAUUACGAUGCCGUGGAGCAGCAGCUCAAGUCCUCGACCAUUGG ...((.((((...((((....))))..))))(((((.(((((...((((..((((.(((....))).))))((((.(((....)))..))))..)))))))))...)))))..))..... ( -39.20) >DroSim_CAF1 196222 120 - 1 GCAGGAGAUCGAGUUCCGCAAGAAGAAGAUCGAGGAGGAGCUCAAGCUGGACAAGAUUGAGAACAAGUUUGCCACCCACUACGAUGCCGUGGAGCAGCAGCUCAAGUCCUCGACCAUUGG ((.((((......))))))........(.(((((((.(((((...(((((.((((.(((....))).)))))).....(((((....))))))))...)))))...))))))).)..... ( -37.40) >DroEre_CAF1 206116 120 - 1 GCAGGAGAUCGAAUUCCGCAAGAAGAAGAUCGGGGAGGAGCUGAAGCUGGAAAAGAUCGAGAACAAGUUUGCCACCCACUACGAUGCCGUGGAGCAGCAGCUUAAGUCCUCGACCAUUGG ...((.((((...((.(....)))...))))(((((.((((((..(((((...((((.........)))).)).....(((((....))))))))..))))))...)))))..))..... ( -35.10) >DroYak_CAF1 203469 120 - 1 GCAGGAGAUCGAGUUCCGCAAAAAGAAGAUCGAGGAGGAGCUGAAGCUGGACAAGAUUGAGAACAAGUUCGCCACCCACUACGAUGCCGUGGAGCAGCAGCUCAAGUCCUCGACCAUUGG ((.((((......))))))........(.(((((((.((((((..(((((......(((....))).....)).....(((((....))))))))..))))))...))))))).)..... ( -38.70) >consensus GCAGGAGAUCGAGUUCCGCAAGAAGAAGAUCGAGGAGGAGCUCAAGCUGGACAAGAUUGAGAACAAGUUUGCCACCCACUACGAUGCCGUGGAGCAGCAGCUCAAGUCCUCGACCAUUGG ((.((((......))))))........(.(((((((.(((((...(((((...(((((.......))))).)).....(((((....))))))))...)))))...))))))).)..... (-33.04 = -32.72 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:19 2006