
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
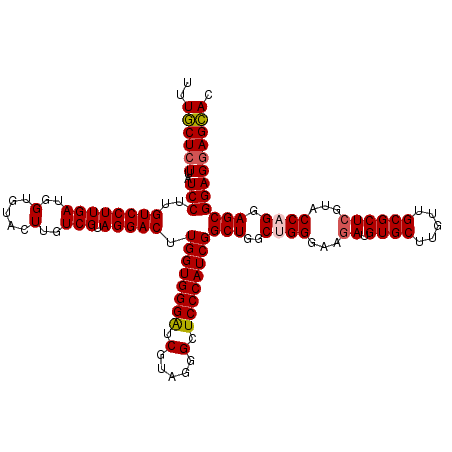
| Location | 23,745,642 – 23,745,802 |
| Length | 160 |
| Max. P | 0.958703 |

| Location | 23,745,642 – 23,745,762 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -44.86 |
| Consensus MFE | -43.14 |
| Energy contribution | -43.46 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
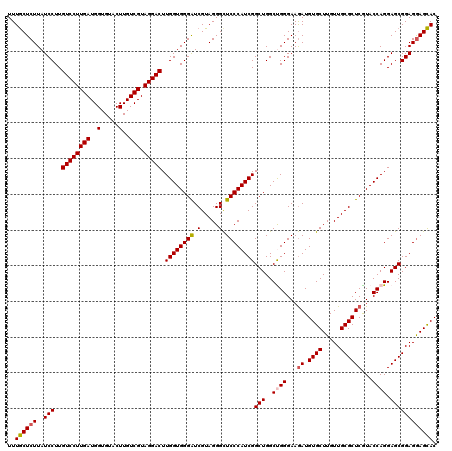
>3R_DroMel_CAF1 23745642 120 + 27905053 UUUACUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGUAC ..((((((..(((..((((((((..(.....)..))).))))).((((((((((....)).))))))))(((..((((...((.((((.....))))))...)))).)))))))))))). ( -45.70) >DroSec_CAF1 195363 120 + 1 UUUGCUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCAUACCAGGAGCGGAGAAGCAC ...(((((.(((((.((((((((..(.....)..))).))))).....)))))...)))))(((((.(.....).)))))....((((((.((.(((((.......))))))).)))))) ( -43.30) >DroSim_CAF1 195342 120 + 1 UUUGCUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCAC ..((((((..(((..((((((((..(.....)..))).))))).((((((((((....)).))))))))(((..((((...((.((((.....))))))...)))).)))))))))))). ( -47.70) >DroEre_CAF1 205242 120 + 1 UUUGCUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCCGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCAC ..(((((((.((((.((((((((..(.....)..))).)))))..(((((((((....)).)))))))((....))((...((.((((.....))))))...))))))....))))))). ( -45.20) >DroYak_CAF1 202595 120 + 1 UUUGCUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGGUCGUAGGGUUCCCAUCGGCUGGCCGGGAAAAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCAC ..((((((..(((..((((((((..(.....)..))).))))).(((((((..(.....)..)))))))(((..((.((.....((((.....)))).....)).)))))))))))))). ( -42.40) >consensus UUUGCUCUUAUCCUUGUCCUUGAUGGUGUACUUGUCGUAGGACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCAC ..((((((..(((..((((((((..(.....)..))).))))).((((((((.(.....).))))))))(((..((((...((.((((.....))))))...)))).)))))))))))). (-43.14 = -43.46 + 0.32)

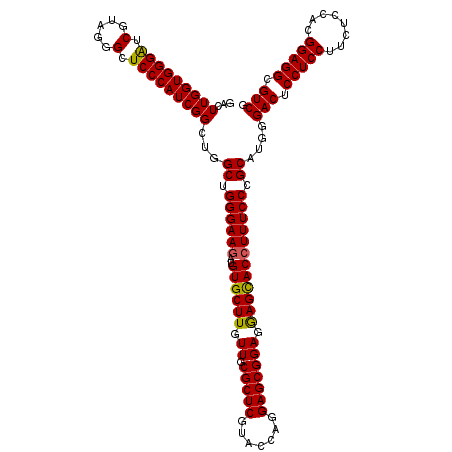
| Location | 23,745,682 – 23,745,802 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -54.72 |
| Consensus MFE | -50.66 |
| Energy contribution | -50.38 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
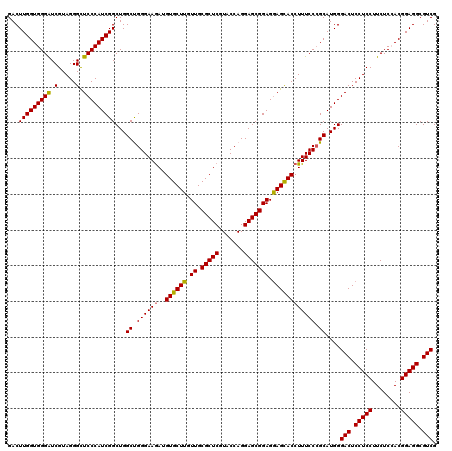
>3R_DroMel_CAF1 23745682 120 + 27905053 GACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGUACCUUUCCGGCAUGGGACUCCUCCUUCUCCACGGAGGCGUCG ...(((((((((((....)).)))))))))...(((((((((..((((((.((.(((((.......))))))).)))))))))))))))....(((.(((((........))))).))). ( -53.70) >DroSec_CAF1 195403 120 + 1 GACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCAUACCAGGAGCGGAGAAGCACCUUUCCCGCAUGGGACUCCUCCUUCUCGACGGAGGCGUCG ...(((((((((((....)).)))))))))...((.((((((..((((((.((.(((((.......))))))).)))))).))))))))....(((.(((((........))))).))). ( -51.40) >DroSim_CAF1 195382 120 + 1 GACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCACCUUUCCCGCAUGGGACUCCUCCUUCUCGACGGAGGCGUCG ..((..((((((....((((((((..((.(((..((((...((.((((.....))))))...)))).))).))))))).))).))))))..))(((.(((((........))))).))). ( -55.90) >DroEre_CAF1 205282 120 + 1 GACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCCGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCACCUUUCCCGCAUGGGACUCCUCCUUCUCCACGGAGGCGUCG ..(((.((((((..(.(((..(((((((((....))((((((..((((((.((.(((((.......))))))).)))))).))))))).))))))..))))..)).)))).)))...... ( -54.80) >DroYak_CAF1 202635 120 + 1 GACUUGGUGGGGUCGUAGGGUUCCCAUCGGCUGGCCGGGAAAAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCACCUUUCCCGCAUGGGACUCCUCCUUCUCCACGGAGGCGUCG ..(((.((((((..(.(((..(((((((((....))((((((..((((((.((.(((((.......))))))).)))))).))))))).))))))..))).)..)))))).)))...... ( -57.80) >consensus GACUUGGUGGGAUCGUAGGGCUCCCAUCGGCUGGCUGGGAAGAUGUGCUUGUUGCGCUCGUACCAGGAGCGGAGGAGCACCUUUCCCGCAUGGGACUCCUCCUUCUCCACGGAGGCGUCG ...(((((((((.(.....).)))))))))...((.((((((..((((((.((.(((((.......))))))).)))))))))))).))....(((.(((((........))))).))). (-50.66 = -50.38 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:15 2006