
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,721,553 – 23,721,681 |
| Length | 128 |
| Max. P | 0.944163 |

| Location | 23,721,553 – 23,721,645 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.19 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
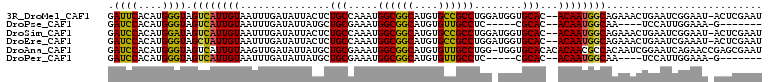
>3R_DroMel_CAF1 23721553 92 + 27905053 ------AGCCACACGCACACUGGCACAUGAUUCACAUGGGUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUG ------.((((.........)))).(((.(((((....(((((.....(((((.....))))).)))))....((((((....))))))))))).))) ( -32.30) >DroPse_CAF1 184574 92 + 1 GGGACUGCUAACCCAUACUCUGGCACA-GAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-----CG ((((((....((((((..((((...))-)).....))))))))))..........(((((.(((((((......))))))))))))....)-----). ( -25.80) >DroSec_CAF1 169159 92 + 1 ------AGCCACACGCACACUGGCACAUGAUCCACAUGGAUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUG ------.((((.........)))).(((.(((((..(((.(((.....(((((.....))))).))))))...((((((....))))))))))).))) ( -32.40) >DroEre_CAF1 178516 92 + 1 ------GAACACACGCACACUGGCACAUGAUCCACAUGGGUAGCUAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUG ------........((......)).(((.(((((....(((((.....(((((.....))))).)))))....((((((....))))))))))).))) ( -31.40) >DroAna_CAF1 175425 80 + 1 ----------CCACCCACAC-------UGAUCCACAUGGGUAGUCAUUGUAAGUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUGG-UGGUG ----------(((((.(((.-------((((((.....))..)))).))).((..(((((.(((((((......))))))))))))..))))-))).. ( -26.70) >DroPer_CAF1 180824 92 + 1 GGAACUGCUAACCCAUACUCUGGCAGA-GAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-----CG (((.((((((..........)))))).-..........((((((((........))))..((((((((......))))))))...))))))-----). ( -26.30) >consensus ______GCCCACACGCACACUGGCACAUGAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUACGCUGCCAAAUGGCGGCAUGUGCCGCCUGG_UGGUG ................(((.((((.....((((....)))).)))).))).......................((((((....))))))......... (-16.65 = -16.65 + 0.00)



| Location | 23,721,575 – 23,721,681 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -21.11 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23721575 106 + 27905053 GAUUCACAUGGGUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUGCAC--ACAAUGGCAGAAACUGAAUCGGAAU-ACUCGAAU ........((((((.((...(((((.....)))))(((((((...((((((....))))))......((....--))..)))))))..........)).)-)))))... ( -33.30) >DroPse_CAF1 184601 90 + 1 GAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-----CGCAC--ACAAUGGCAA----UCCAUUGGAAA-G------- ..((((.((((((.((((((((...........(((((((......)))))))((((......-----.))))--)))))))).)----)))))))))..-.------- ( -34.90) >DroSim_CAF1 169004 106 + 1 GAUCCACAUGGAUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUGCAC--ACAAUGGCAGAAACUGAAUCGGAAU-ACUCGAAU .((((....))))..((...(((.((((((.....(((((((...((((((....))))))......((....--))..)))))))......)))))).)-))..)).. ( -33.00) >DroEre_CAF1 178538 106 + 1 GAUCCACAUGGGUAGCUAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUGGAUGGUGCAC--ACAAUGGCAGAAACUGAAUCGAAAU-ACUCGAAU .((((....)))).......(((.((((((.....(((((((...((((((....))))))......((....--))..)))))))......)))))).)-))...... ( -32.70) >DroAna_CAF1 175436 108 + 1 GAUCCACAUGGGUAGUCAUUGUAAGUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUGG-UGGUGCACACACAACGCCACAAUCGGAAUCAGAACCGAGCGAAU .((((....))))..((.((((..((((.....(((((((......)))))))((((((((...-.)))).)))).))))...))))((((........))))..)).. ( -31.10) >DroPer_CAF1 180851 90 + 1 GAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-----CGCAC--ACAAUGGCAA----UCCAUUGGAAA-G------- ..((((.((((((.((((((((...........(((((((......)))))))((((......-----.))))--)))))))).)----)))))))))..-.------- ( -34.90) >consensus GAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUACGCUGCCAAAUGGCGGCAUGUGCCGCCUGG_UGGUGCAC__ACAAUGGCAGAAACUGAAUCGGAAU_ACUCGAAU .((((....)))).((((((((...............(((.....((((((....))))))........)))...)))))))).......................... (-21.11 = -20.42 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:07 2006