
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,659,064 – 23,659,158 |
| Length | 94 |
| Max. P | 0.871235 |

| Location | 23,659,064 – 23,659,158 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -14.09 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
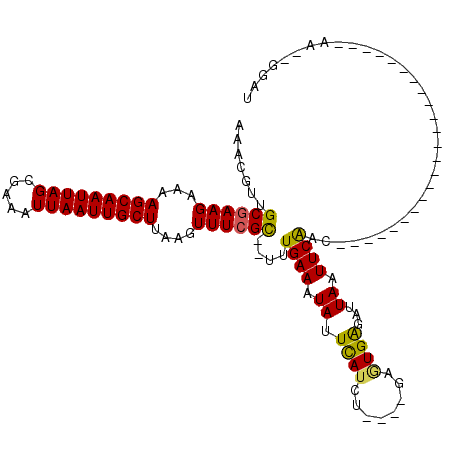
>3R_DroMel_CAF1 23659064 94 + 27905053 AAACGUGGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUCAAGUUUCGC--UUUGAAAUAUUCAUCC------GUGAGAUUAAUUCGCCGGCCG----------------ACAA--AGCU ...(((((((((....(((((((((.....)))))))))..((((((((--..(((.....)))...------))))))))..)))))))..))----------------....--.... ( -24.90) >DroVir_CAF1 166492 94 + 1 AAACGUUGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUUAAGUUUCGC-UUUUGAAAUAUUUAUCUGCCCGAGUGAGAUUAAUUCAAC-----------------------AA--GGAU .......((((((..((((((((((.....))))))))))...))))))-(((((....(((.((((((....)).)))).)))....)-----------------------))--)).. ( -19.20) >DroGri_CAF1 156976 94 + 1 AAACGUUGCAAAGAAAAGCAAUUAGCGAAAUUAAUUGCUGAAGUUUCGC-UUUUGAAAUAUUUAUCUGCCCGAAUGAGAUUAAUUCAAC-----------------------AA--GCAU ((((.((((........))))(((((((......))))))).)))).((-(((((((.((...((((.........)))))).))))).-----------------------))--)).. ( -16.60) >DroSec_CAF1 121000 94 + 1 AAACGUGGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUCAAGUUUCGC--UUUGAAAUAUUCAUCA------GUGAGAUUAAUUCGCCGGCCG----------------ACAA--AGCU ...(((((((((....(((((((((.....)))))))))..((((((((--(.(((.....)))..)------))))))))..)))))))..))----------------....--.... ( -26.10) >DroWil_CAF1 138689 114 + 1 AAACGGAGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUUAAGUUUCGU--UUUGAAAUAUUCAUCU----GAGUGGGAUUAAUUCAACAGUCGGGGUCGGGGCCAACUGAAGAAGGGCU ...((((((((((..((((((((((.....))))))))))...))))))--))))...(((((....----))))).......(((..((((...(((....))).))))..)))..... ( -31.00) >DroMoj_CAF1 179747 95 + 1 AAACGUUGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUUAAGUUUCGCCCUUUGAAAUAUUUAUCUGCUCGACUGACAUUAAUUCAAC-----------------------AA--GGAU .......((((((..((((((((((.....))))))))))...))))))((((((((.((....((.........))...)).))))..-----------------------))--)).. ( -19.20) >consensus AAACGUUGCGAAGAAAAGCAAUUAGCGAAAUUAAUUGCUUAAGUUUCGC__UUUGAAAUAUUCAUCU____GAGUGAGAUUAAUUCAAC_______________________AA__GGAU .......((((((...(((((((((.....)))))))))....))))))....((((.((.((((........))))...)).))))................................. (-14.09 = -13.57 + -0.52)


| Location | 23,659,064 – 23,659,158 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -13.04 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
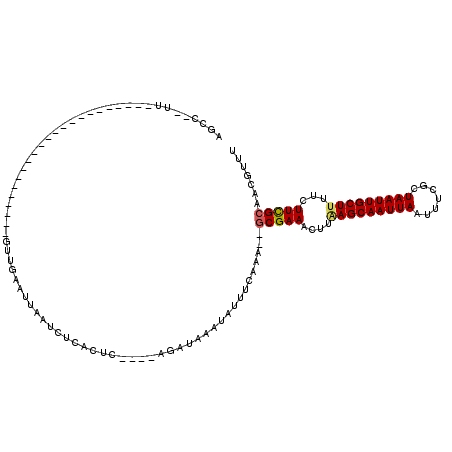
>3R_DroMel_CAF1 23659064 94 - 27905053 AGCU--UUGU----------------CGGCCGGCGAAUUAAUCUCAC------GGAUGAAUAUUUCAAA--GCGAAACUUGAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCCACGUUU .(((--....----------------.))).((((((...(((....------.)))(((........(--((((((.((((....)))))))))))........)))))))))...... ( -21.59) >DroVir_CAF1 166492 94 - 1 AUCC--UU-----------------------GUUGAAUUAAUCUCACUCGGGCAGAUAAAUAUUUCAAAA-GCGAAACUUAAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCAACGUUU ....--..-----------------------.(((((.(((((((......).))))...)).)))))..-(((((....(((((((((.......)))))))))...)))))....... ( -17.80) >DroGri_CAF1 156976 94 - 1 AUGC--UU-----------------------GUUGAAUUAAUCUCAUUCGGGCAGAUAAAUAUUUCAAAA-GCGAAACUUCAGCAAUUAAUUUCGCUAAUUGCUUUUCUUUGCAACGUUU .(((--((-----------------------(.(((.......)))..))))))................-(((((.....((((((((.......))))))))....)))))....... ( -18.20) >DroSec_CAF1 121000 94 - 1 AGCU--UUGU----------------CGGCCGGCGAAUUAAUCUCAC------UGAUGAAUAUUUCAAA--GCGAAACUUGAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCCACGUUU .(((--....----------------.))).((((((...(((....------.)))(((........(--((((((.((((....)))))))))))........)))))))))...... ( -21.79) >DroWil_CAF1 138689 114 - 1 AGCCCUUCUUCAGUUGGCCCCGACCCCGACUGUUGAAUUAAUCCCACUC----AGAUGAAUAUUUCAAA--ACGAAACUUAAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCUCCGUUU (((..(((..(((((((........)))))))..)))............----.((.(((..((((...--..))))...(((((((((.......)))))))))))).)))))...... ( -21.70) >DroMoj_CAF1 179747 95 - 1 AUCC--UU-----------------------GUUGAAUUAAUGUCAGUCGAGCAGAUAAAUAUUUCAAAGGGCGAAACUUAAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCAACGUUU ..((--((-----------------------..((((...((((..(((.....)))..))))))))))))(((((....(((((((((.......)))))))))...)))))....... ( -21.40) >consensus AGCC__UU_______________________GUUGAAUUAAUCUCACUC____AGAUAAAUAUUUCAAA__GCGAAACUUAAGCAAUUAAUUUCGCUAAUUGCUUUUCUUCGCAACGUUU .......................................................................(((((....(((((((((.......)))))))))...)))))....... (-13.04 = -13.07 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:56 2006