
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,644,794 – 23,644,899 |
| Length | 105 |
| Max. P | 0.910456 |

| Location | 23,644,794 – 23,644,899 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
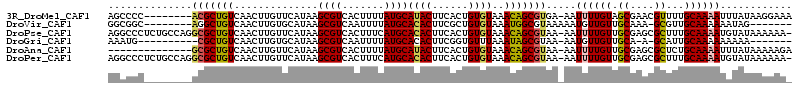
>3R_DroMel_CAF1 23644794 105 + 27905053 AGCCCC--------ACGCUGUCAACUUGUUCAUAAGCGUCACUUUUAUGCAUACUUCACUGUGUAAACAGCGUGA-AAUUUUGUAGCGAACGUUUUGCAAAAUUUAUAAGGAAA ...(((--------(((((((..............((((.......))))((((......))))..))))))))(-((((((((((........)))))))))))....))... ( -25.20) >DroVir_CAF1 146721 98 + 1 GGCGGC--------AGGCUGUCAACUUGUGCAUAAGCGUCAAUUUUAUGCACACUUCGCUGUGUAAAUGGCGUAAAAAUGUUGUUGCAAA-GCGUUGCAAAAAAUAG------- (((((.--------...)))))....((((((((((.......))))))))))...((((.((((.(..((((....))))..))))).)-))).............------- ( -28.60) >DroPse_CAF1 109431 112 + 1 AGGCCCUCUGCCAGGCGCUGUCAACUUGUUCAUAAGCGUCACUUUCAUGCACACUUCACUGUGUAAACAGCGUAA-AAUUUUGUUGCGAGCGCUUUGCAAAAUGUAUAAAAAA- .(((.....)))..(((((((..............((((.......))))((((......))))..)))))))..-.(((((((.((....))...)))))))..........- ( -27.00) >DroGri_CAF1 139161 94 + 1 AAAUG----------CGCUGUCAACUUGUGCAUAAGCGUCAAUUUUAUGCACACUUCGGUGUUUAAAUAGCGUAA-AAUGUUGUUGCA-A-GCAUUGCAAAAAAAAA------- ...((----------((((((.(((..(((((((((.......)))))))))((....)))))...)))))))).-((((((......-)-)))))...........------- ( -25.30) >DroAna_CAF1 107756 99 + 1 --------------GCGCUGUCAACUUGUUCAUAAGCGUCACUUUUAUGCAUACUUCACUGUGUAAACAGCGUAA-AAUUUUGUUGCGAGCGCUCUGCAAAAUUUAUAAAAAGA --------------(((((((..............((((.......))))((((......))))..))))))).(-((((((((.((....))...)))))))))......... ( -22.60) >DroPer_CAF1 107736 112 + 1 AGGCCCUCUGCCAGGCGCUGUCAACUUGUUCAUAAGCGUCACUUUCAUGCACACUUCACUGUGUAAACAGCGUAA-AAUUUUGUUGCGAGCGCUUUGCAAAAUGUAUAAAAAA- .(((.....)))..(((((((..............((((.......))))((((......))))..)))))))..-.(((((((.((....))...)))))))..........- ( -27.00) >consensus AGCCCC________GCGCUGUCAACUUGUUCAUAAGCGUCACUUUUAUGCACACUUCACUGUGUAAACAGCGUAA_AAUUUUGUUGCGAACGCUUUGCAAAAUAUAUAAAAAA_ ..............(((((((..............((((.......))))((((......))))..))))))).....((((((.((....))...))))))............ (-16.43 = -16.43 + 0.00)
| Location | 23,644,794 – 23,644,899 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -15.68 |
| Energy contribution | -17.27 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
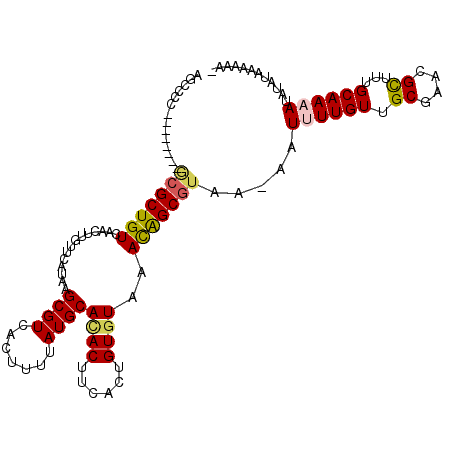
>3R_DroMel_CAF1 23644794 105 - 27905053 UUUCCUUAUAAAUUUUGCAAAACGUUCGCUACAAAAUU-UCACGCUGUUUACACAGUGAAGUAUGCAUAAAAGUGACGCUUAUGAACAAGUUGACAGCGU--------GGGGCU ..(((....((((((((..............)))))))-)(((((((((.((...((.((((...(((....)))..))))....))..)).))))))))--------)))).. ( -22.44) >DroVir_CAF1 146721 98 - 1 -------CUAUUUUUUGCAACGC-UUUGCAACAACAUUUUUACGCCAUUUACACAGCGAAGUGUGCAUAAAAUUGACGCUUAUGCACAAGUUGACAGCCU--------GCCGCC -------.........(((..((-(...((((..........(((..........)))...(((((((((.........))))))))).))))..))).)--------)).... ( -21.10) >DroPse_CAF1 109431 112 - 1 -UUUUUUAUACAUUUUGCAAAGCGCUCGCAACAAAAUU-UUACGCUGUUUACACAGUGAAGUGUGCAUGAAAGUGACGCUUAUGAACAAGUUGACAGCGCCUGGCAGAGGGCCU -...........((((((...(((((..((((......-...((((((....))))))((((((.(((....)))))))))........))))..)))))...))))))..... ( -35.20) >DroGri_CAF1 139161 94 - 1 -------UUUUUUUUUGCAAUGC-U-UGCAACAACAUU-UUACGCUAUUUAAACACCGAAGUGUGCAUAAAAUUGACGCUUAUGCACAAGUUGACAGCG----------CAUUU -------...........(((((-(-((((((......-...((............))...(((((((((.........))))))))).)))).))).)----------)))). ( -18.30) >DroAna_CAF1 107756 99 - 1 UCUUUUUAUAAAUUUUGCAGAGCGCUCGCAACAAAAUU-UUACGCUGUUUACACAGUGAAGUAUGCAUAAAAGUGACGCUUAUGAACAAGUUGACAGCGC-------------- .....................(((((..((((......-...((((((....))))))((((...(((....)))..))))........))))..)))))-------------- ( -21.30) >DroPer_CAF1 107736 112 - 1 -UUUUUUAUACAUUUUGCAAAGCGCUCGCAACAAAAUU-UUACGCUGUUUACACAGUGAAGUGUGCAUGAAAGUGACGCUUAUGAACAAGUUGACAGCGCCUGGCAGAGGGCCU -...........((((((...(((((..((((......-...((((((....))))))((((((.(((....)))))))))........))))..)))))...))))))..... ( -35.20) >consensus _UUUUUUAUAAAUUUUGCAAAGCGCUCGCAACAAAAUU_UUACGCUGUUUACACAGUGAAGUGUGCAUAAAAGUGACGCUUAUGAACAAGUUGACAGCGC________GGGCCU .....................(((((..((((..........((((((....))))))((((((.(((....)))))))))........))))..))))).............. (-15.68 = -17.27 + 1.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:47 2006