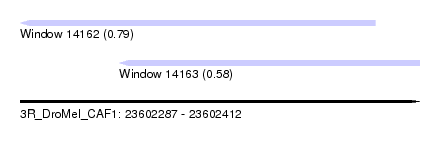
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,602,287 – 23,602,412 |
| Length | 125 |
| Max. P | 0.787579 |

| Location | 23,602,287 – 23,602,398 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.98 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23602287 111 - 27905053 UUUUAUUUUGAGUAUUUGUACUUAAACAUAUUUAAAAUGAAUUUCUUUGCUUUCGAUGCCAAUAAUUGGGAGAAGGCAAAAGAACUUGGGUAAAUAUGGUCCAAUCCUCGU ......((((((((....))))))))((((((((...(.((.((((((((((((....(((.....)))..)))))).)))))).)).).))))))))............. ( -26.20) >DroSec_CAF1 63533 111 - 1 UUUUAUUUUGAGUAUUUGUGCUUUAACAUAUUUUAAAGGAAUUUCUUUGCUUUCGAUGGCAAUAAUUGGGAGAAGGCAAAGAAACUUGGGUAAAUAAGGUCCAAUCCUCGU .........(((...(((..(((((...((((...(((...(((((((((((((.................)))))))))))))))).))))..)))))..)))..))).. ( -24.23) >DroEre_CAF1 65322 111 - 1 UAUUAUCUUGAGUAUUUGUCUCUAAAUAUAUUUUAAAGGAAUUUCCUUGCUUUCGAUGCCAAUAAUUGGAAGAAGGCAAGGGAACUUGGGUAAAUACGGUCCAAACCUCGU ...........(((((((.(((((((.....))))......(((((((((((((....(((.....)))..)))))))))))))...))))))))))(((....))).... ( -29.90) >DroYak_CAF1 69198 100 - 1 UUUUAUCUUAAGUGUUUG-----------AAUUGAGAUGAAGUUCUUUGCUUUCGAUGUCAAUAAUUGGCAGAAGCCAAAGGAUCUUUAGUAAAUAAAGUGCAAACAUCGU (((((((((((.......-----------..)))))))))))...........((((((......(((((....))))).(.(.(((((.....)))))).)..)))))). ( -21.80) >consensus UUUUAUCUUGAGUAUUUGU_CUUAAACAUAUUUUAAAGGAAUUUCUUUGCUUUCGAUGCCAAUAAUUGGGAGAAGGCAAAGGAACUUGGGUAAAUAAGGUCCAAACCUCGU .((((((((((((((............))))))))))))))(((((((((((((....(((.....)))..)))))))))))))........................... (-17.54 = -18.98 + 1.44)


| Location | 23,602,318 – 23,602,412 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -11.12 |
| Energy contribution | -12.57 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23602318 94 - 27905053 -UAUAUUUGCAA-UCAUUUUAUUUUGAGUAUUUGUACUUAAACAUAUUUAAAAUGAAUUUCUUUGCUUUCGAUGCCAAUAAUUGGGAGAAGGCAAA -...........-((((((((.((((((((....))))))))......))))))))......((((((((....(((.....)))..)))))))). ( -20.60) >DroSec_CAF1 63564 95 - 1 -UAUAUUUACAACUCAUUUUAUUUUGAGUAUUUGUGCUUUAACAUAUUUUAAAGGAAUUUCUUUGCUUUCGAUGGCAAUAAUUGGGAGAAGGCAAA -..........(((((........))))).....((((((.........((((((....))))))((..((((.......))))..)))))))).. ( -17.90) >DroYak_CAF1 69229 85 - 1 AGAUAUUUCCAAGGCAUUUUAUCUUAAGUGUUUG-----------AAUUGAGAUGAAGUUCUUUGCUUUCGAUGUCAAUAAUUGGCAGAAGCCAAA .((((((..(((((.((((((((((((.......-----------..)))))))))))).))))).....)))))).....(((((....))))). ( -24.20) >consensus _UAUAUUUACAA_UCAUUUUAUUUUGAGUAUUUGU_CUU_AACAUAUUUAAAAUGAAUUUCUUUGCUUUCGAUGCCAAUAAUUGGGAGAAGGCAAA .................((((((((((((((............)))))))))))))).....((((((((....(((.....)))..)))))))). (-11.12 = -12.57 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:30 2006