
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,601,558 – 23,601,751 |
| Length | 193 |
| Max. P | 0.941047 |

| Location | 23,601,558 – 23,601,656 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
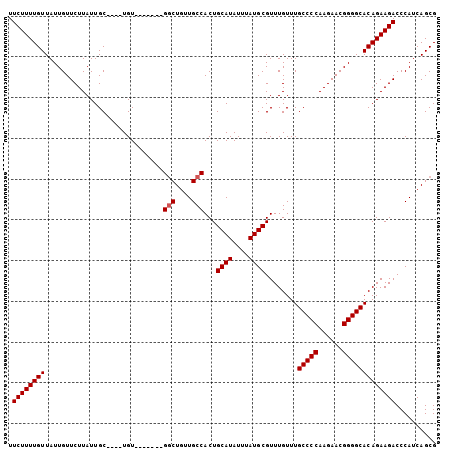
>3R_DroMel_CAF1 23601558 98 + 27905053 UUCUUUUGUUAUUGUUCUUAUUGCUGGCUGU-------GGCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG ......................((((((.((-------(((....))))).))......(((.((((...((((((......))))))....)))).))))))). ( -33.30) >DroSec_CAF1 62812 94 + 1 UUCUUUUGUUAUUGUUCUUAUUGC----UGU-------GGCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG .((((((((....((......(((----.((-------(((....))))).))).......))........(((((......))))))))))))).......... ( -31.62) >DroSim_CAF1 64614 94 + 1 UUCUUUUGUUAUUGUUCUUAUUGC----UGU-------GGCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG .((((((((....((......(((----.((-------(((....))))).))).......))........(((((......))))))))))))).......... ( -31.62) >DroEre_CAF1 64603 94 + 1 UUCUUUUGUUAUUGUUCUUAUUGC----UGU-------GGCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG .((((((((....((......(((----.((-------(((....))))).))).......))........(((((......))))))))))))).......... ( -31.62) >DroYak_CAF1 68471 101 + 1 UUCUUUUGUUAUUGUUCUUAUUGC----UGUUGUGGCUGUCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG ......................((----((..(((((........))))).((((....))))((((...((((((......))))))....))))....)))). ( -30.50) >consensus UUCUUUUGUUAUUGUUCUUAUUGC____UGU_______GGCUGUUGCCACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCG .((((((((.............................(((....)))...((((....))))........(((((......))))))))))))).......... (-24.26 = -24.46 + 0.20)


| Location | 23,601,558 – 23,601,656 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
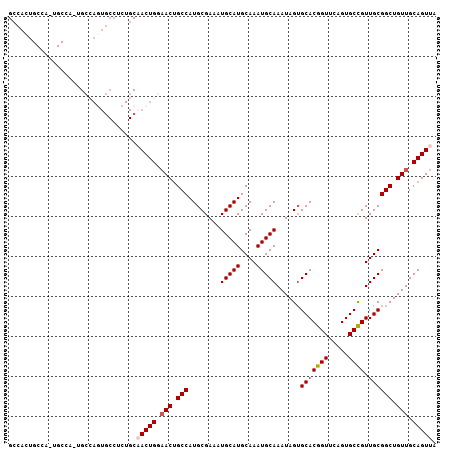
>3R_DroMel_CAF1 23601558 98 - 27905053 CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGCC-------ACAGCCAGCAAUAAGAACAAUAACAAAAGAA .(((((((.((......(((((((....))))))).....)).)))......((.(((((....)))-------)).))))))...................... ( -33.00) >DroSec_CAF1 62812 94 - 1 CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGCC-------ACA----GCAAUAAGAACAAUAACAAAAGAA .......(((.(......).)))(((((((..((........((((....)))).(((((....)))-------)).----))..)))))))............. ( -29.20) >DroSim_CAF1 64614 94 - 1 CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGCC-------ACA----GCAAUAAGAACAAUAACAAAAGAA .......(((.(......).)))(((((((..((........((((....)))).(((((....)))-------)).----))..)))))))............. ( -29.20) >DroEre_CAF1 64603 94 - 1 CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGCC-------ACA----GCAAUAAGAACAAUAACAAAAGAA .......(((.(......).)))(((((((..((........((((....)))).(((((....)))-------)).----))..)))))))............. ( -29.20) >DroYak_CAF1 68471 101 - 1 CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGACAGCCACAACA----GCAAUAAGAACAAUAACAAAAGAA .((((.(((((..(((((((((((....))))))........((((....)))).......)))))..))).)).))----))...................... ( -34.00) >consensus CGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGUGGCAACAGCC_______ACA____GCAAUAAGAACAAUAACAAAAGAA .(((((((.((......(((((((....))))))).....)).))).....(((....))).))))....................................... (-23.00 = -23.20 + 0.20)



| Location | 23,601,584 – 23,601,691 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -32.57 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23601584 107 + 27905053 GCUGU-------GGCUGUUGCC-ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACGGCACUGAACCGUGCA (.(((-------(((((((((.-...((((....))))((((...((((((......))))))....))))..............)))))))))))).).((((......)))). ( -44.60) >DroSec_CAF1 62836 105 + 1 --UGU-------GGCUGUUGCC-ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACGGCACUGAACCGUGCA --(((-------(((((((((.-...((((....))))((((...((((((......))))))....))))..............))))))))))))...((((......)))). ( -44.30) >DroSim_CAF1 64638 105 + 1 --UGU-------GGCUGUUGCC-ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACGGCACUGAACCGUGCA --(((-------(((((((((.-...((((....))))((((...((((((......))))))....))))..............))))))))))))...((((......)))). ( -44.30) >DroEre_CAF1 64627 105 + 1 --UGU-------GGCUGUUGCC-ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACAGCACUGAACCGUGCU --(((-------(((((((((.-...((((....))))((((...((((((......))))))....))))..............))))))))))))..(((((......))))) ( -44.50) >DroYak_CAF1 68495 112 + 1 --UGUUGUGGCUGUCUGUUGCC-ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACGGCACUGAACCGUGCA --.(((((((((((..((((((-...((((....))))((((...((((((......))))))....))))......).)))))....))))))))))).((((......)))). ( -46.60) >DroPer_CAF1 63649 91 + 1 --UGU-------UGCUGCUGCUGACUGCAUAUUUAUGCGUUUGUUUGCC---------------AGGCAACGCUCUUGCGACACAGCAACAGCAAUGGCGGCAUUGAACUGUGAA --.((-------(((((.(((((...((((....))))(((((.....)---------------))))..(((....)))...))))).))))))).((((.......))))... ( -29.50) >consensus __UGU_______GGCUGUUGCC_ACUGCAUAUUUAUGCGUUUGUUUGCCCCAAGAACGGGGCACAGAAGACCCAUCAGCGUAACUGCAACAGCCGCAACGGCACUGAACCGUGCA ............(((((((((.....((((....))))((((...((((((......))))))....))))..............)))))))))......((((......)))). (-32.57 = -33.43 + 0.86)



| Location | 23,601,584 – 23,601,691 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -42.39 |
| Consensus MFE | -33.83 |
| Energy contribution | -34.93 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23601584 107 - 27905053 UGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU-GGCAACAGCC-------ACAGC .((((......)))).((((.(((((((((.....((((((((.((......(((((((....))))))).....)).))).......))))-))))))))))-------.)))) ( -45.71) >DroSec_CAF1 62836 105 - 1 UGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU-GGCAACAGCC-------ACA-- .(((((((.....)))).)))(((((((((.....((((((((.((......(((((((....))))))).....)).))).......))))-))))))))))-------...-- ( -45.51) >DroSim_CAF1 64638 105 - 1 UGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU-GGCAACAGCC-------ACA-- .(((((((.....)))).)))(((((((((.....((((((((.((......(((((((....))))))).....)).))).......))))-))))))))))-------...-- ( -45.51) >DroEre_CAF1 64627 105 - 1 AGCACGGUUCAGUGCUGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU-GGCAACAGCC-------ACA-- .(((((((.....)))).)))(((((((((.....((((((((.((......(((((((....))))))).....)).))).......))))-))))))))))-------...-- ( -43.51) >DroYak_CAF1 68495 112 - 1 UGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU-GGCAACAGACAGCCACAACA-- .((((......)))).((((.(((((((...(((((((..(((.((......(((((((....))))))).....)).)))......)).))-)))....))))))).)))).-- ( -46.00) >DroPer_CAF1 63649 91 - 1 UUCACAGUUCAAUGCCGCCAUUGCUGUUGCUGUGUCGCAAGAGCGUUGCCU---------------GGCAAACAAACGCAUAAAUAUGCAGUCAGCAGCAGCA-------ACA-- ....................((((((((((((..((....))((((.....---------------.((........))......))))...)))))))))))-------)..-- ( -28.10) >consensus UGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUACGCUGAUGGGUCUUCUGUGCCCCGUUCUUGGGGCAAACAAACGCAUAAAUAUGCAGU_GGCAACAGCC_______ACA__ .(((((((.....)))).)))(((((((((......................(((((((....))))))).......((((....)))).....)))))))))............ (-33.83 = -34.93 + 1.10)



| Location | 23,601,656 – 23,601,751 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -24.36 |
| Energy contribution | -25.96 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23601656 95 + 27905053 UAACUGCAACAGCCGCAACGGCACUGAACCGUGCACUAUUUGCAUUUGCAUGCAUUUCGCAUGGCAGUUCCAGUUGCAGAGGCACUGGC------------AGUGGC ..(((((....((((((((((.((((..((((((......(((((....)))))....)))))))))).)).)))))...)))....))------------)))... ( -36.90) >DroSec_CAF1 62906 95 + 1 UAACUGCAACAGCCGCAACGGCACUGAACCGUGCACUAUUUGCAUUUGCAUGCAUUUCGCAUGGCAGUUCCAGUUGCAGAGGCACUGGC------------AAUGGC ...(((((((.((((...))))((((..((((((......(((((....)))))....))))))))))....)))))))..((....))------------...... ( -32.70) >DroSim_CAF1 64708 107 + 1 UAACUGCAACAGCCGCAACGGCACUGAACCGUGCACUAUUUGCAUUUGCAUGCAUUUCGCAUGGCAGUUCCAGUUGCAGAGGCACUGGCACUGGCAAUGGCAGUGGC ...(((((((.((((...))))((((..((((((......(((((....)))))....))))))))))....))))))).(.(((((.((.......)).))))).) ( -41.00) >DroEre_CAF1 64697 107 + 1 UAACUGCAACAGCCGCAACAGCACUGAACCGUGCUCUAUUUGCAUUUGCAUGCAUUUCGCAUGGCAGUUCCAGUGGCAAAGGCAGUUCCAGUGGCAGUGGCAGUGGC ..(((((.((.(((((.....((((((((.((((.......)))).(((((((.....)))).)))))).)))))((....)).......))))).)).)))))... ( -36.90) >DroYak_CAF1 68572 95 + 1 UAACUGCAACAGCCGCAACGGCACUGAACCGUGCACUAUUUGCAUUUGCAUGCAUUUCGCAUGGCUGUGGCAGUGG------------CAGUGGCAGUGGCAGUGGC ..(((((.((.(((((...(.(((((..((((((......(((((....)))))....))))))......))))).------------).))))).)).)))))... ( -36.90) >consensus UAACUGCAACAGCCGCAACGGCACUGAACCGUGCACUAUUUGCAUUUGCAUGCAUUUCGCAUGGCAGUUCCAGUUGCAGAGGCACUGGCA_UGGCA_UGGCAGUGGC ...(((((((.((((...))))((((..((((((......(((((....)))))....))))))))))....)))))))............................ (-24.36 = -25.96 + 1.60)



| Location | 23,601,656 – 23,601,751 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23601656 95 - 27905053 GCCACU------------GCCAGUGCCUCUGCAACUGGAACUGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGUGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUA ...(((------------((....(((...(((((.((.((((((.((((...(((((....))))).....)))).))..)))).))))))))))....))))).. ( -34.60) >DroSec_CAF1 62906 95 - 1 GCCAUU------------GCCAGUGCCUCUGCAACUGGAACUGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGUGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUA ......------------.(((((((....)).)))))((((((.........(((((....)))))(((((((((((((.....)))).))).)))))))))))). ( -33.80) >DroSim_CAF1 64708 107 - 1 GCCACUGCCAUUGCCAGUGCCAGUGCCUCUGCAACUGGAACUGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGUGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUA ((.((.(((.((((((((.(((((((....)).))))).)))).(((((.....))))).....)))).....(((((((.....)))).)))))).)).))..... ( -39.60) >DroEre_CAF1 64697 107 - 1 GCCACUGCCACUGCCACUGGAACUGCCUUUGCCACUGGAACUGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGAGCACGGUUCAGUGCUGUUGCGGCUGUUGCAGUUA ...(((((.((.(((((((((..(((.(((((...(((.....))).))))).(((((....)))))......)))...))))))((....))))).)).))))).. ( -35.20) >DroYak_CAF1 68572 95 - 1 GCCACUGCCACUGCCACUG------------CCACUGCCACAGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGUGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUA ((....((....))....)------------).(((((.((((((..((....(((((....)))))....))(((((((.....)))).))))))))).))))).. ( -34.10) >consensus GCCACUGCCA_UGCCA_UGCCAGUGCCUCUGCAACUGGAACUGCCAUGCGAAAUGCAUGCAAAUGCAAAUAGUGCACGGUUCAGUGCCGUUGCGGCUGUUGCAGUUA ................................(((((.(((.(((........(((((....)))))......(((((((.....)))).)))))).))).))))). (-27.50 = -27.94 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:28 2006