
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,596,903 – 23,597,028 |
| Length | 125 |
| Max. P | 0.988230 |

| Location | 23,596,903 – 23,596,998 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -15.98 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
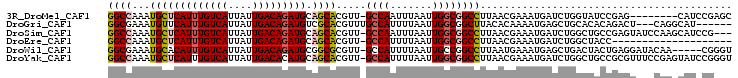
>3R_DroMel_CAF1 23596903 95 + 27905053 GCUCGGAUG--------CUCGGAUACCAGAUCAUUUCGUUAAGGCCGCCAAUUAAAUUGGC-AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC ...((((((--------.((........)).)).))))....((((((((((...))))))-...(((((.(((.(((((....))))).))))))))..)))) ( -29.60) >DroGri_CAF1 83190 95 + 1 ------AUGCCUG---AGUCUGUGUGCAGCUCAUUUUGUGUAAGCCGCCAAUUAAAAUGGCAAACGUGCGACAUCUGUCAAUAAUGACAAAUGAACAUUUCGCC ------.((((((---((.(((....)))))))....(((.....)))..........)))).....(((((((.(((((....))))).)))......)))). ( -22.40) >DroSim_CAF1 59940 100 + 1 ---CGGAUGCUUGGAUACUCGGCAGCCAGAUCAUUUCGUUAAGGCCGCCAAUUAAAUUGGC-AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC ---.((.((((.((...)).)))).))...............((((((((((...))))))-...(((((.(((.(((((....))))).))))))))..)))) ( -32.90) >DroEre_CAF1 60205 83 + 1 --------------------GGUAGCCAGAUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGC-AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC --------------------((..(((.((.....)).....)))..)).........(((-...(((((.(((.(((((....))))).))))))))...))) ( -26.30) >DroWil_CAF1 68818 98 + 1 ACCCG-----UUGUAUCCUCAGUAGUCAGCUCAUUUCAUUAAGGCCGGCAAUUAAAAUGGC-AACGCGCCGCAUCUGUCAAUAAUGACAAAUGUGCAUUUCGCC ....(-----((((......(((.(((.(((...........))).))).)))......))-)))(((.(((((.(((((....))))).))))).....))). ( -22.10) >DroYak_CAF1 63557 103 + 1 ACCCGGAUACUCGGAAACGCGGCAGCCAGAUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGC-AACGUGCUGCAUGUGUCAAUAAUGACAAAUGAGCAUUUGGCC ..((((((.((((....)(((((((((.((.....)).....))).((((.......))))-....))))))...(((((....)))))...))).)))))).. ( -32.40) >consensus ___CG_AUG_UUG___ACUCGGUAGCCAGAUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGC_AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC ..........................................((((((((.......))))....(((((.(((.(((((....))))).))))))))..)))) (-15.98 = -17.32 + 1.33)


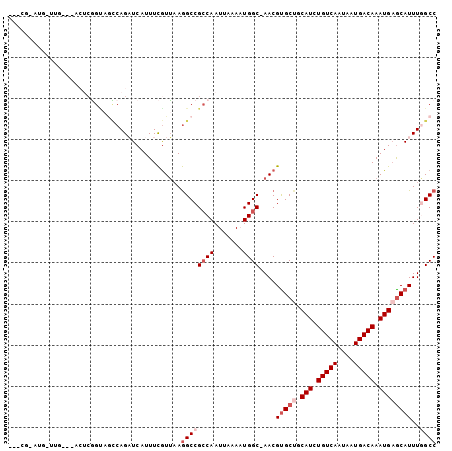
| Location | 23,596,903 – 23,596,998 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
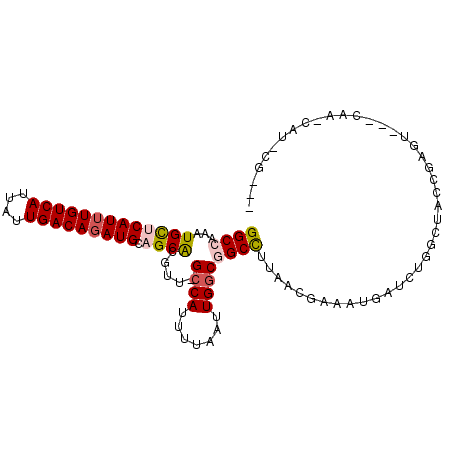
>3R_DroMel_CAF1 23596903 95 - 27905053 GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU-GCCAAUUUAAUUGGCGGCCUUAACGAAAUGAUCUGGUAUCCGAG--------CAUCCGAGC ((((...(((((((((((((....))))))))).))))....-((((((...))))))))))....((..(((.((.((...)))).--------))).))... ( -32.40) >DroGri_CAF1 83190 95 - 1 GGCGAAAUGUUCAUUUGUCAUUAUUGACAGAUGUCGCACGUUUGCCAUUUUAAUUGGCGGCUUACACAAAAUGAGCUGCACACAGACU---CAGGCAU------ ((((((.(((.(((((((((....)))))))))..)))..))))))........((((((((((.......)))))))).))......---.......------ ( -31.10) >DroSim_CAF1 59940 100 - 1 GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU-GCCAAUUUAAUUGGCGGCCUUAACGAAAUGAUCUGGCUGCCGAGUAUCCAAGCAUCCG--- (((.(..(((((((((((((....))))))))).))))..).-))).......(((((((((...............)))))))))...............--- ( -36.86) >DroEre_CAF1 60205 83 - 1 GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU-GCCAUUUUAAUUGGCGGCCUUAACGAAAUGAUCUGGCUACC-------------------- (((((..(((((((((((((....))))))))).)))).(((-((((.......)))))))...............)))))...-------------------- ( -32.30) >DroWil_CAF1 68818 98 - 1 GGCGAAAUGCACAUUUGUCAUUAUUGACAGAUGCGGCGCGUU-GCCAUUUUAAUUGCCGGCCUUAAUGAAAUGAGCUGACUACUGAGGAUACAA-----CGGGU (((((..(((.(((((((((....)))))))))..)))..))-)))..........(((.(((((..................)))))......-----))).. ( -28.17) >DroYak_CAF1 63557 103 - 1 GGCCAAAUGCUCAUUUGUCAUUAUUGACACAUGCAGCACGUU-GCCAUUUUAAUUGGCGGCCUUAACGAAAUGAUCUGGCUGCCGCGUUUCCGAGUAUCCGGGU ((((...(((((((.(((((....))))).))).))))....-((((.......))))))))...........((((((.(((((......)).))).)))))) ( -35.80) >consensus GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU_GCCAUUUUAAUUGGCGGCCUUAACGAAAUGAUCUGGCUACCGAGU___CAA_CAU_CG___ ((((...(((((((((((((....))))))))).)))).....((((.......)))))))).......................................... (-23.75 = -24.33 + 0.59)

| Location | 23,596,934 – 23,597,028 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23596934 94 + 27905053 UAAGGCCGCCAAUUAAAUUGGC-------AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC---------ACUGGAGUCGGGCUUCAAAGGGCGAAUCUG ...((((((((((...))))))-------...(((((.(((.(((((....))))).))))))))..))))---------..(((((.....)))))............. ( -32.10) >DroVir_CAF1 88835 93 + 1 UUGAGCCGCCAAUUAAAAUGGCA------AACGUGCGACAUCUGUCAAUAAUGACAAAUGAACAUUUCGCC---------AUUAAAGCCAGGCC--CAGAAUCAUCUCUG ....(((((((.......)))).------...(.(((((((.(((((....))))).)))......)))))---------..........))).--((((......)))) ( -22.00) >DroPse_CAF1 60196 109 + 1 UUAAGCCGCCAAUUAAAAUGGCAACGGGCAACGUGCAACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCAUUCAGGCGAUCGAAGCGGCGACUCAAAG-GCGCCUCUC ....(((((((.......))))...((.(((.((((..(((.(((((....))))).))).))))))).)).....)))......((.((((.((....)-))))).)). ( -35.10) >DroEre_CAF1 60224 94 + 1 UAAGGCCGCCAAUUAAAAUGGC-------AACGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC---------ACUGAAGUCGGGCUUCAAAGGGCGAGUCGG ...(((((((........((((-------...(((((.(((.(((((....))))).))))))))...)))---------).(((((.....)))))...)))).))).. ( -36.20) >DroYak_CAF1 63596 94 + 1 UAAGGCCGCCAAUUAAAAUGGC-------AACGUGCUGCAUGUGUCAAUAAUGACAAAUGAGCAUUUGGCC---------ACUGAAGUCGGGCUUCAAAGGGCGUAUCUG ......((((........((((-------...(((((.(((.(((((....))))).))))))))...)))---------).(((((.....)))))...))))...... ( -31.70) >DroPer_CAF1 58519 109 + 1 UUAAGCCGCCAAUUAAAAUGGCAACGGGCAACGUGCAACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCAUUCAGGCGAUCGAAGCGGCGACUCAAAG-GCGCCUCUC ....(((((((.......))))...((.(((.((((..(((.(((((....))))).))).))))))).)).....)))......((.((((.((....)-))))).)). ( -35.10) >consensus UAAAGCCGCCAAUUAAAAUGGC_______AACGUGCUACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCC_________ACUGAAGCCGGGCCUCAAAGGGCGACUCUG ....(((((((.......))))..........((((..(((.(((((....))))).))).))))..)))........................................ (-15.28 = -15.62 + 0.33)



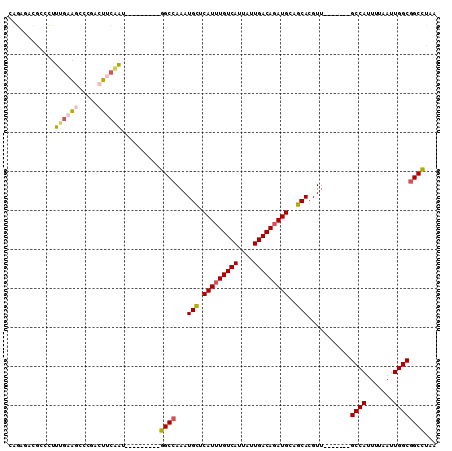
| Location | 23,596,934 – 23,597,028 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23596934 94 - 27905053 CAGAUUCGCCCUUUGAAGCCCGACUCCAGU---------GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU-------GCCAAUUUAAUUGGCGGCCUUA ..((.(((..((....))..))).))....---------((((...(((((((((((((....))))))))).))))....-------((((((...))))))))))... ( -33.10) >DroVir_CAF1 88835 93 - 1 CAGAGAUGAUUCUG--GGCCUGGCUUUAAU---------GGCGAAAUGUUCAUUUGUCAUUAUUGACAGAUGUCGCACGUU------UGCCAUUUUAAUUGGCGGCUCAA ............((--((((..(((..(((---------((((((.(((.(((((((((....)))))))))..)))..))------)))))))......))))))))). ( -34.00) >DroPse_CAF1 60196 109 - 1 GAGAGGCGC-CUUUGAGUCGCCGCUUCGAUCGCCUGAAUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGUUGCACGUUGCCCGUUGCCAUUUUAAUUGGCGGCUUAA ..((((((.-(........).))))))....(((..(((((.(((.(((.(((((((((....)))))))))..)))..))).)))))((((.......))))))).... ( -39.80) >DroEre_CAF1 60224 94 - 1 CCGACUCGCCCUUUGAAGCCCGACUUCAGU---------GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU-------GCCAUUUUAAUUGGCGGCCUUA .......(((..((((((.....)))))).---------)))....(((((((((((((....))))))))).)))).(((-------((((.......))))))).... ( -34.70) >DroYak_CAF1 63596 94 - 1 CAGAUACGCCCUUUGAAGCCCGACUUCAGU---------GGCCAAAUGCUCAUUUGUCAUUAUUGACACAUGCAGCACGUU-------GCCAUUUUAAUUGGCGGCCUUA .......(((..((((((.....)))))).---------)))....(((((((.(((((....))))).))).)))).(((-------((((.......))))))).... ( -31.10) >DroPer_CAF1 58519 109 - 1 GAGAGGCGC-CUUUGAGUCGCCGCUUCGAUCGCCUGAAUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGUUGCACGUUGCCCGUUGCCAUUUUAAUUGGCGGCUUAA ..((((((.-(........).))))))....(((..(((((.(((.(((.(((((((((....)))))))))..)))..))).)))))((((.......))))))).... ( -39.80) >consensus CAGAGACGCCCUUUGAAGCCCGACUUCAAU_________GGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACGUU_______GCCAUUUUAAUUGGCGGCCUAA ............((((((.....))))))..........((((...(((.(((((((((....)))))))))..)))...........((((.......))))))))... (-24.89 = -25.17 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:23 2006