
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,484,437 – 23,484,557 |
| Length | 120 |
| Max. P | 0.500000 |

| Location | 23,484,437 – 23,484,557 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -30.57 |
| Energy contribution | -29.99 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23484437 120 + 27905053 CUGCUCCUUCUCCUUCUUGACCUGUGAGUAGAUGGGCAGGAACUCAUCCAGCUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCGCCCAAUUUCUCGAUCAGCGCCAAGGUGGGGUUCAA ((((.((.(((.((((.......).))).))).))))))((((((...((((..((......))..))))(((..((((((((.(..(........)..).))))))))))))))))).. ( -35.40) >DroVir_CAF1 99337 120 + 1 CUGCUCCUUCUCCUUCUUCACUUGCGAGUAGAUGGGCAGGAAUUCGUCCAAUUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCACCCAUUUUCUCGAUCAGAGCCAGGGUGGGGUUCAA .((..(((..((((..(((.....((((.(((((((..(((.....)))....................(((((....)))))..))))))).))))....)))..))))..)))..)). ( -34.00) >DroGri_CAF1 97392 120 + 1 CUGCUCCUUCUCCUUCUUCACUUGCGAGUAGAUGGGCAGGAAUUCGUCCAACUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCGCCCAUUUUCUCGAUCAGCGCCAAGGUGGGGUUCAA ..(((((....((((...(.((..((((.((((((((.(((.....)))(((..((......))..)))(((((....))))).)))))))).))))...)).)..)))).))))).... ( -38.00) >DroWil_CAF1 79699 120 + 1 CUGUUCCUUCUCCUUCUUAACCUGUGAGUAGAUGGGCAGGAACUCAUCCAACUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCACCCAUUUUCUCGAUGAGAGCCAAGGUGGGGUUCAA .((..((((..((((((((.....((((.(((((((..(((.....)))(((..((......))..)))(((((....)))))..))))))).)))).)))))....))).))))..)). ( -31.40) >DroMoj_CAF1 108978 120 + 1 CUGCUCCUUCUCCUUCUUCACUUGCGAGUAGAUGGGCAGGAAUUCCUCCAAUUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCGCCCAUUUUCUCGAUCAAAGCGAGGGUGGGGUUCAA .........((((.(((((.(((.((((.((((((((.(((.....)))....................(((((....))))).)))))))).))))....))).))))).))))..... ( -40.90) >DroPer_CAF1 82924 120 + 1 CUGCUCCUUCUCCUUCUUGACCUGUGAGUAAAUGGGCAGGAAUUCGUCCAUCUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCACCCAUUUUCUCGAUCAGAGCCAAGGUGGGGUUCAA .........((((.(((((.((((((((.((((((((((((.........)))))..............(((((....)))))..))))))).))))..))).).))))).))))..... ( -33.60) >consensus CUGCUCCUUCUCCUUCUUCACCUGCGAGUAGAUGGGCAGGAAUUCGUCCAACUUGAUCUUCUUCUCGUUGCGCUUCUUGGUGCCACCCAUUUUCUCGAUCAGAGCCAAGGUGGGGUUCAA ..(((((....((((.........((((.(((((((..(((.....)))....................(((((....)))))..))))))).)))).........)))).))))).... (-30.57 = -29.99 + -0.58)

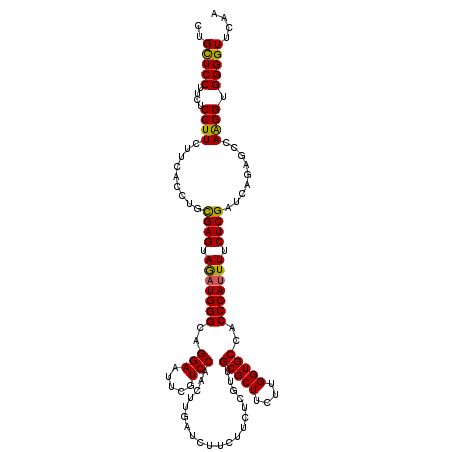
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:50 2006