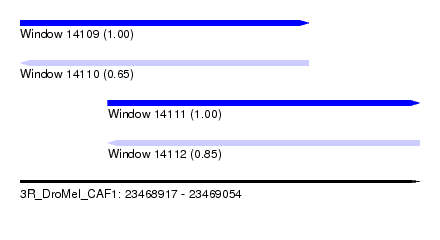
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,468,917 – 23,469,054 |
| Length | 137 |
| Max. P | 0.995829 |

| Location | 23,468,917 – 23,469,016 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23468917 99 + 27905053 AUGGUAGCUACGGAACUUCAAGAGGUUUGCAAUCACUUCAACAAAUGUCUAAAGGUAUGC-----------AGCUACUUUCUAUUUUUCAAAGCAUACUUUUAGACACUU .((((.((....((((((....)))))))).))))..........(((((((((((((((-----------.....................)))))))))))))))... ( -25.40) >DroSec_CAF1 68351 110 + 1 AAGGUAGCUAACCAACUUCAAGAGGUUUGCAAUCACUUCAAAAAAUGUCUAAAAGUAAGCAGCGAACUAGCAGCUACUUUCUAUUUUCCAAAGCAUACUUUUAGACACUU ..(((.((.((((..........)))).)).)))...........(((((((((((((((.((......)).))).((((.........))))..))))))))))))... ( -25.70) >DroEre_CAF1 58994 108 + 1 A-GGUAGCUACGCUACUACAAGGGGUUUGCAAUCACUUGCA-ACAUGUCUGAAAGUAUGCAGCGAAUUACAAGCUACUUUCUAUUUUUCAUAGCAUACUUUUGGACACUU .-((((((...)))))).........((((((....)))))-)..((((..((((((((((((.........)))......(((.....))))))))))))..))))... ( -31.10) >consensus A_GGUAGCUACGCAACUUCAAGAGGUUUGCAAUCACUUCAA_AAAUGUCUAAAAGUAUGCAGCGAA_UA__AGCUACUUUCUAUUUUUCAAAGCAUACUUUUAGACACUU ..(((.((.....(((((....))))).)).)))...........(((((((((((((((................................)))))))))))))))... (-20.04 = -20.38 + 0.34)


| Location | 23,468,917 – 23,469,016 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -16.48 |
| Energy contribution | -20.37 |
| Covariance contribution | 3.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23468917 99 - 27905053 AAGUGUCUAAAAGUAUGCUUUGAAAAAUAGAAAGUAGCU-----------GCAUACCUUUAGACAUUUGUUGAAGUGAUUGCAAACCUCUUGAAGUUCCGUAGCUACCAU (((((((((((.((((((.....................-----------)))))).)))))))))))......(((.((((.(((........)))..)))).)))... ( -21.30) >DroSec_CAF1 68351 110 - 1 AAGUGUCUAAAAGUAUGCUUUGGAAAAUAGAAAGUAGCUGCUAGUUCGCUGCUUACUUUUAGACAUUUUUUGAAGUGAUUGCAAACCUCUUGAAGUUGGUUAGCUACCUU (((((((((((((((.((..((((...(((..........))).))))..)).)))))))))))))))............((.((((..........)))).))...... ( -28.20) >DroEre_CAF1 58994 108 - 1 AAGUGUCCAAAAGUAUGCUAUGAAAAAUAGAAAGUAGCUUGUAAUUCGCUGCAUACUUUCAGACAUGU-UGCAAGUGAUUGCAAACCCCUUGUAGUAGCGUAGCUACC-U ..(((((..(((((((((..((((..((((........))))..))))..)))))))))..))))).(-(((((....))))))..........(((((...))))).-. ( -31.20) >consensus AAGUGUCUAAAAGUAUGCUUUGAAAAAUAGAAAGUAGCU__UA_UUCGCUGCAUACUUUUAGACAUUU_UUGAAGUGAUUGCAAACCUCUUGAAGUUGCGUAGCUACC_U ((((((((((((((((((..((((....((.......)).....))))..))))))))))))))))))......((((((.(((.....))).))))))........... (-16.48 = -20.37 + 3.89)

| Location | 23,468,947 – 23,469,054 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -25.03 |
| Energy contribution | -25.15 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23468947 107 + 27905053 AAUCACUUCAACAAAUGUCUAAAGGUAUGC-----------AGCUACUUUCUAUUUUUCAAAGCAUACUUUUAGACACUUCAUAGAUGGUUUCAAAUCUCUAAAGAUUUCGGUGGCUC ..(((((........(((((((((((((((-----------.....................)))))))))))))))(((.(.((((........)))).).))).....)))))... ( -25.30) >DroSec_CAF1 68381 118 + 1 AAUCACUUCAAAAAAUGUCUAAAAGUAAGCAGCGAACUAGCAGCUACUUUCUAUUUUCCAAAGCAUACUUUUAGACACUUUAUAGAUGGUUUCAAGUCUCCAAAGAUUUCGGUGGCUC ..(((((........(((((((((((((((.((......)).))).((((.........))))..))))))))))))((((..((((........))))..)))).....)))))... ( -25.90) >DroEre_CAF1 59023 117 + 1 AAUCACUUGCA-ACAUGUCUGAAAGUAUGCAGCGAAUUACAAGCUACUUUCUAUUUUUCAUAGCAUACUUUUGGACACUUCGCAGAUGCUUUCAAAUCUCUGUAGAUUUCGGUGGCUC ......((((.-...((((..((((((((((((.........)))......(((.....))))))))))))..))))....))))..(((.((((((((....)))))).)).))).. ( -30.70) >consensus AAUCACUUCAA_AAAUGUCUAAAAGUAUGCAGCGAA_UA__AGCUACUUUCUAUUUUUCAAAGCAUACUUUUAGACACUUCAUAGAUGGUUUCAAAUCUCUAAAGAUUUCGGUGGCUC ..(((((........(((((((((((((((................................)))))))))))))))(((((.((((........)))).))))).....)))))... (-25.03 = -25.15 + 0.12)


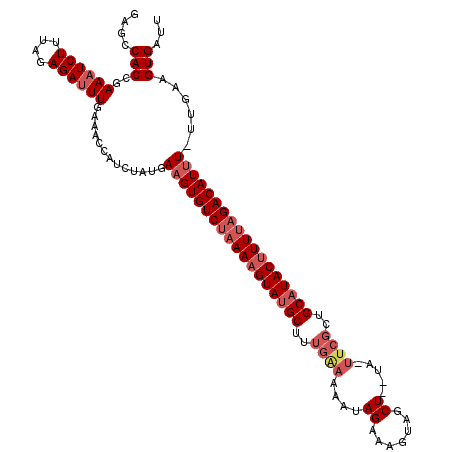
| Location | 23,468,947 – 23,469,054 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -20.38 |
| Energy contribution | -23.60 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23468947 107 - 27905053 GAGCCACCGAAAUCUUUAGAGAUUUGAAACCAUCUAUGAAGUGUCUAAAAGUAUGCUUUGAAAAAUAGAAAGUAGCU-----------GCAUACCUUUAGACAUUUGUUGAAGUGAUU ....(((..((((((....)))))).............(((((((((((.((((((.....................-----------)))))).)))))))))))......)))... ( -24.10) >DroSec_CAF1 68381 118 - 1 GAGCCACCGAAAUCUUUGGAGACUUGAAACCAUCUAUAAAGUGUCUAAAAGUAUGCUUUGGAAAAUAGAAAGUAGCUGCUAGUUCGCUGCUUACUUUUAGACAUUUUUUGAAGUGAUU ....(((.....((...(....)..)).....((...((((((((((((((((.((..((((...(((..........))).))))..)).))))))))))))))))..)).)))... ( -29.50) >DroEre_CAF1 59023 117 - 1 GAGCCACCGAAAUCUACAGAGAUUUGAAAGCAUCUGCGAAGUGUCCAAAAGUAUGCUAUGAAAAAUAGAAAGUAGCUUGUAAUUCGCUGCAUACUUUCAGACAUGU-UGCAAGUGAUU .........((((((....)))))).....((..(((((.(((((..(((((((((..((((..((((........))))..))))..)))))))))..))))).)-))))..))... ( -32.20) >consensus GAGCCACCGAAAUCUUUAGAGAUUUGAAACCAUCUAUGAAGUGUCUAAAAGUAUGCUUUGAAAAAUAGAAAGUAGCU__UA_UUCGCUGCAUACUUUUAGACAUUU_UUGAAGUGAUU ....(((..((((((....)))))).............((((((((((((((((((..((((....((.......)).....))))..))))))))))))))))))......)))... (-20.38 = -23.60 + 3.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:43 2006