
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,446,113 – 23,446,214 |
| Length | 101 |
| Max. P | 0.957103 |

| Location | 23,446,113 – 23,446,214 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
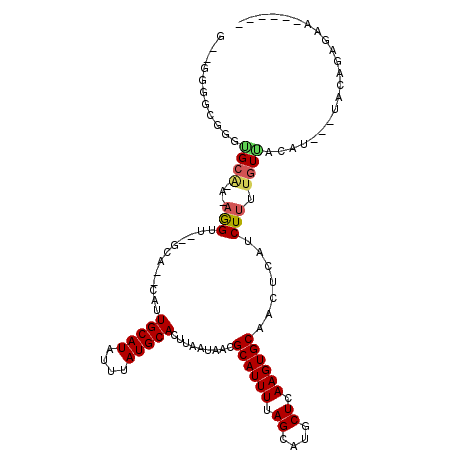
>3R_DroMel_CAF1 23446113 101 + 27905053 ------UCCUGCGAA---AUGUAACAAAAGAUGAGAUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUG--CUC--AACCUUU-UGCACUCGGCACCAC ------.........---...........(.((((((((((((((..((((......))))..)))))))))).....(((((..--...--......)-)))))))).)..... ( -24.10) >DroVir_CAF1 49332 98 + 1 ------U---AUGUG---CUGUAACAAAAGAUGAGUUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUG--CGCGCAGCCA-UACGCUCGCUCUAU--C ------(---(((.(---((((..............(((((((((..((((......))))..))))))))).......((....--.)))))))))-))............--. ( -26.70) >DroPse_CAF1 41879 108 + 1 UUUUCUCUCUCUGUA---AUGUUACAAAAGAUGAGUUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUGUGUGC--AACCU-U-UGCACCUGCCCCUGC ....((((((.((((---....))))..))).))).(((((((((..((((......))))..))))))))).......(((..(.((((--(....-.-)))))))))...... ( -27.20) >DroGri_CAF1 46263 101 + 1 --------CUCAUUCUGUAUGUAACAAAAGAUGAGUUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUG--AGCGGAGCUU-U-UGCCACCGCCCU--C --------.((((..(((((((..((.....))...(((((((((..((((......))))..)))))))))...))))))))))--)((((.((..-.-.))..))))...--. ( -29.10) >DroWil_CAF1 36398 96 + 1 ------AUG-----U---CUGUAACAAAAGAUGAGCUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUG--UGC--AACCUCU-GCCCCUCAACACACA ------..(-----(---((........))))(((.(((((((((..((((......))))..)))))))))......(((....--.))--)......-....)))........ ( -22.30) >DroPer_CAF1 41548 107 + 1 -UUUCUCUCUCUGUA---AUGUUACAAAAGAUGAGUUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUGUGUGC--AACCU-U-UGCACCUGCCCCUGC -...((((((.((((---....))))..))).))).(((((((((..((((......))))..))))))))).......(((..(.((((--(....-.-)))))))))...... ( -27.20) >consensus ______CUCUCUGUA___AUGUAACAAAAGAUGAGUUGCACUUGAGCAUGCUAAAAUGCGUUAUUAAGUGCAUAAAUAUGCAAUG__UGC__AACCU_U_UGCACCCGCCAC__C ....................................(((((((((..((((......))))..)))))))))........................................... (-15.70 = -15.70 + 0.00)



| Location | 23,446,113 – 23,446,214 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -11.49 |
| Energy contribution | -11.52 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23446113 101 - 27905053 GUGGUGCCGAGUGCA-AAAGGUU--GAG--CAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAUCUCAUCUUUUGUUACAU---UUCGCAGGA------ ....(((.(((((((-((((((.--(((--...(((((....))))).........((((((.((....)).))))))..)))))))))))...)))---)).)))...------ ( -27.00) >DroVir_CAF1 49332 98 - 1 G--AUAGAGCGAGCGUA-UGGCUGCGCG--CAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAACUCAUCUUUUGUUACAG---CACAU---A------ .--...(((((.(((((-....))))).--...(((((....))))).........((......)).)))))..((((....((.....)).....)---)))..---.------ ( -23.00) >DroPse_CAF1 41879 108 - 1 GCAGGGGCAGGUGCA-A-AGGUU--GCACACAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAACUCAUCUUUUGUAACAU---UACAGAGAGAGAAAA (((.(((((((((((-.-(((((--(((.....)))).)))).)))))))......((......))..))))...)))..(((.(((((.(((....---))))))))))).... ( -30.90) >DroGri_CAF1 46263 101 - 1 G--AGGGCGGUGGCA-A-AAGCUCCGCU--CAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAACUCAUCUUUUGUUACAUACAGAAUGAG-------- .--.((((((.(((.-.-..))))))))--)..(((((....))))).........((((((.((....)).))))))..(((((..(((((.....))))))))))-------- ( -30.90) >DroWil_CAF1 36398 96 - 1 UGUGUGUUGAGGGGC-AGAGGUU--GCA--CAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAGCUCAUCUUUUGUUACAG---A-----CAU------ (((.(((.(((((..-.(((.((--(((--(..(((((....))))).........((((......))))....))))))))).)))))....))).---)-----)).------ ( -26.50) >DroPer_CAF1 41548 107 - 1 GCAGGGGCAGGUGCA-A-AGGUU--GCACACAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAACUCAUCUUUUGUAACAU---UACAGAGAGAGAAA- (((.(((((((((((-.-(((((--(((.....)))).)))).)))))))......((......))..))))...)))..(((.(((((.(((....---)))))))))))...- ( -30.90) >consensus G__GGGGCGGGUGCA_A_AGGUU__GCA__CAUUGCAUAUUUAUGCACUUAAUAACGCAUUUUAGCAUGCUCAAGUGCAACUCAUCUUUUGUUACAU___UACAGAGAA______ ...........((((...(((............(((((....))))).........((((((.((....)).)))))).......))).))))...................... (-11.49 = -11.52 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:36 2006