
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,436,159 – 23,436,258 |
| Length | 99 |
| Max. P | 0.572498 |

| Location | 23,436,159 – 23,436,258 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
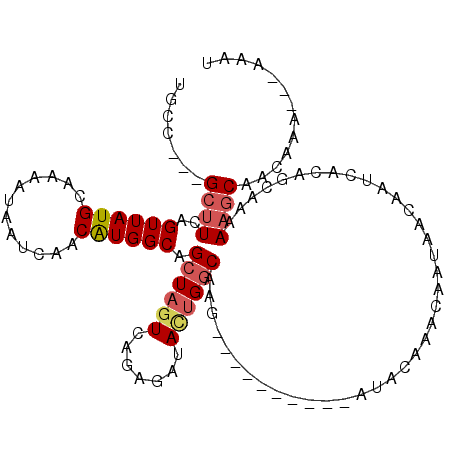
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -9.51 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
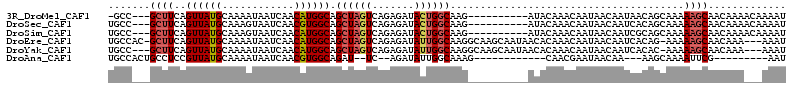
>3R_DroMel_CAF1 23436159 99 - 27905053 -GCC---GCUUCAGUUAUGCAAAAUAAUCAACAUGGCAGCUAGUCAGAGAUACUGGCAAG----------AUACAAACAAUAACAAUAACAGCAAAAAGCAACAAAACAAAAU -(((---(.....(((((.....))))).....)))).((((((.......))))))...----------.....................((.....))............. ( -14.40) >DroSec_CAF1 28815 100 - 1 UGCC---GCUUCAGUUAUGCAAAGUAAUCAACGUGGCAGCUAGUCAGAGAUACUGGCAAG----------AUACAAACAAUAACAAUCACAGCAAAAAGCAACAAAACAAAAU ((((---((.(..(((((.....)))))..).))))))(((.(((((.....)))))..(----------((.............)))..))).................... ( -18.22) >DroSim_CAF1 26819 100 - 1 UGCC---GCUUCAGUUAUGCAAAGUAAUCAACAUGGCAGCUAGUCAGAGAUACUGGCAAG----------AUACAAACAAUAACAAUCGCAGCAAAAAGCAACAAAACAAAAU (((.---((....(((((((...((.....))...)).((((((.......))))))...----------.........)))))....)).)))................... ( -16.00) >DroEre_CAF1 25513 108 - 1 UGCCAC-GCUUCAGUUAUGCAAAAUAAUCAACAUGGCAGCUAGUCAGAGAUAUUGGCAAGGCAAGCAAUAACACAAACAAUAACAAUCACAG-AAAAAGCAACAAA---AAAU ((((..-......((((((............)))))).((((((.......))))))..)))).............................-.............---.... ( -14.40) >DroYak_CAF1 26336 106 - 1 UGCC---GCUUCAGUUAUGCAAAAUAAUCAACAUGGCAGCUAGUCAGAGAUAUUGGCAAGGCAAGCAAUAACACAAACAAUAACAAUCACAC-AAAAAGCAACAAA---AAAU ((((---......((((((............)))))).((((((.......))))))..)))).............................-.............---.... ( -14.40) >DroAna_CAF1 27876 85 - 1 UGCCACUGCCUCCGUUAUGCAAAAUAAUCAACGUGGCAGAU--UC--AGAUAUUGGCAAAG------------CAACGAAUAACAA---AAGCAAAAUUCG---------AAU ((((((((((..((((((........)).)))).)))))..--..--......)))))...------------...(((((.....---.......)))))---------... ( -17.10) >consensus UGCC___GCUUCAGUUAUGCAAAAUAAUCAACAUGGCAGCUAGUCAGAGAUACUGGCAAG__________AUACAAACAAUAACAAUCACAGCAAAAAGCAACAAA___AAAU .......((((..((((((............)))))).((((((.......)))))).......................................))))............. ( -9.51 = -9.95 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:31 2006