
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,405,046 – 23,405,163 |
| Length | 117 |
| Max. P | 0.960941 |

| Location | 23,405,046 – 23,405,139 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.54 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23405046 93 + 27905053 A--GUGAUCUUUUUUUCAUCGCCACUUAAUUGCACCCUAGUCACGUUCGGUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGAUUA .--((((........))))..((((......((((((.((.((((((((((..(....).....))-)))))))))).))).)))...)))).... ( -29.90) >DroSec_CAF1 19514 93 + 1 A--GUGAUCUUGUUUUCAUCGUCACUUAAUUGCAACCUAGUCACGUUCGAUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUA (--(((((..((....))..)))))).....(((((((((.((((((((....(....).......-)))))))).)))))))))........... ( -28.20) >DroSim_CAF1 19735 93 + 1 A--GUGAUCUUGUUUUCAUCGCCACUUAAUUGCAACCUAGUCACGUUCGAUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUA .--((((........))))..((((......(((((((((.((((((((....(....).......-)))))))).)))))))))...)))).... ( -28.50) >DroEre_CAF1 22888 91 + 1 AUGAUGAUCUUGUUUUCAUCGCCACUUAAUUGCACCCUAGUCACGUUCGAUUUGGGAACCUAACGC-UGGGCGUGCUUAGGUUGCCUUGUGG---- ..(((((........))))).((((......(((.(((((.((((((((..((((....))))...-)))))))).))))).)))...))))---- ( -30.60) >DroYak_CAF1 20862 92 + 1 AUGAUGAUCUGGUUUUCAUCGCCACUUAAUUGCACCCUAGUCACGUUCGAUUUGGGAACUUUACGCCUGGGCGUGCCUAGGUUGCCUUGUGG---- ..(((((........))))).((((......(((.(((((.(((((((.......)))).....((....))))).))))).)))...))))---- ( -29.70) >consensus A__GUGAUCUUGUUUUCAUCGCCACUUAAUUGCACCCUAGUCACGUUCGAUUUGGGAACUUUACGC_UGGGCGUGCUUGGGUUGCCUUGUGG__UA ...((((........))))..((((......(((.(((((.((((((((...((.........))..)))))))).))))).)))...)))).... (-24.34 = -23.54 + -0.80)



| Location | 23,405,046 – 23,405,139 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23405046 93 - 27905053 UAAUCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAACCGAACGUGACUAGGGUGCAAUUAAGUGGCGAUGAAAAAAAGAUCAC--U ....((((...(((.(((.(((((((...-)).....((((.......))))))).)).))))))......))))..................--. ( -20.20) >DroSec_CAF1 19514 93 - 1 UAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAAUCGAACGUGACUAGGUUGCAAUUAAGUGACGAUGAAAACAAGAUCAC--U ...........((((((..(((((((...-)).....((((.......))))))).)).)))))).....((((((..((....)).).))))--) ( -19.00) >DroSim_CAF1 19735 93 - 1 UAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAAUCGAACGUGACUAGGUUGCAAUUAAGUGGCGAUGAAAACAAGAUCAC--U ..(((.((...((((((..(((((((...-)).....((((.......))))))).)).))))))......))))).................--. ( -20.80) >DroEre_CAF1 22888 91 - 1 ----CCACAAGGCAACCUAAGCACGCCCA-GCGUUAGGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUAAGUGGCGAUGAAAACAAGAUCAUCAU ----((((...((.......)).(((((.-..((((.((((.......)))).))))..))))).......)))).(((((........))))).. ( -25.80) >DroYak_CAF1 20862 92 - 1 ----CCACAAGGCAACCUAGGCACGCCCAGGCGUAAAGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUAAGUGGCGAUGAAAACCAGAUCAUCAU ----((((...(((.(((((.(((((....)).....((((.......))))))).))))).)))......)))).(((((........))))).. ( -29.50) >consensus UA__CCACAAGGCAACCCAAGCACGCCCA_GCGUAAAGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUAAGUGGCGAUGAAAACAAGAUCAC__U ....((((...(((.(((.(((((((....)).....((((.......))))))).)).))))))......))))..................... (-17.58 = -17.78 + 0.20)

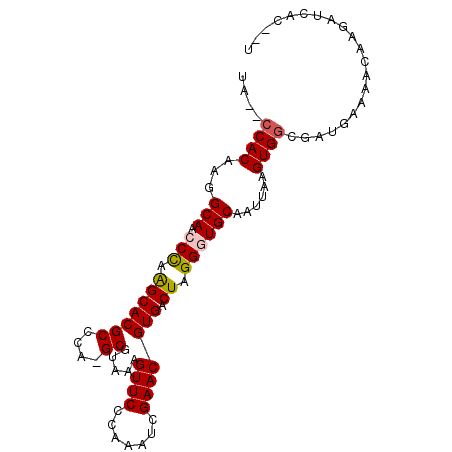
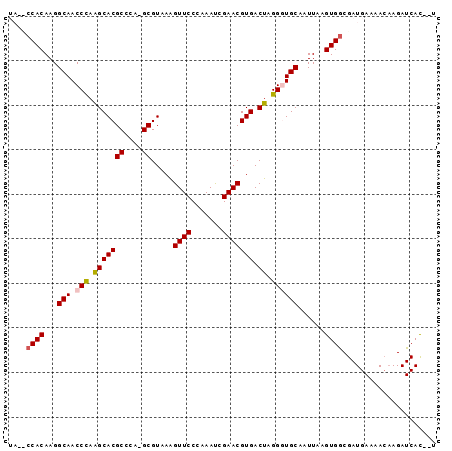
| Location | 23,405,070 – 23,405,163 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23405070 93 + 27905053 UAAUUGCACCCUAGUCACGUUCGGUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGAUUACAUGGAUUAUGGAUUAUAAUUAAA (((((((((((.((.((((((((((..(....).....))-)))))))))).))).))).(..((.....))..)......)))))........ ( -26.20) >DroSec_CAF1 19538 93 + 1 UAAUUGCAACCUAGUCACGUUCGAUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUAUAUGGAUUAUGGAUUAUAAUUAAA .....(((((((((.((((((((....(....).......-)))))))).))))))))).((((((.(((((.....))))).))))))..... ( -27.00) >DroSim_CAF1 19759 93 + 1 UAAUUGCAACCUAGUCACGUUCGAUUUGGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUAUAUGGAUUAUGGAUUAUAAUUAAA .....(((((((((.((((((((....(....).......-)))))))).))))))))).((((((.(((((.....))))).))))))..... ( -27.00) >DroEre_CAF1 22914 80 + 1 UAAUUGCACCCUAGUCACGUUCGAUUUGGGAACCUAACGC-UGGGCGUGCUUAGGUUGCCUUGUGG---------AUUAUGG----CAAUAGUG .....((((((((((...((((.......)))).....))-)))).))))....((((((......---------.....))----)))).... ( -23.70) >DroYak_CAF1 20888 81 + 1 UAAUUGCACCCUAGUCACGUUCGAUUUGGGAACUUUACGCCUGGGCGUGCCUAGGUUGCCUUGUGG---------AUUAUGG----CAAUUGAA (((((((.......(((((..(((((((((.....(((((....))))))))))))))...)))))---------......)----)))))).. ( -26.22) >consensus UAAUUGCACCCUAGUCACGUUCGAUUUGGGAACUUUACGC_UGGGCGUGCUUGGGUUGCCUUGUGG__UA_AUGGAUUAUGGAUUAUAAUUAAA .....(((.(((((.((((((((...((.........))..)))))))).))))).)))................................... (-18.92 = -18.52 + -0.40)



| Location | 23,405,070 – 23,405,163 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.685000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23405070 93 - 27905053 UUUAAUUAUAAUCCAUAAUCCAUGUAAUCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAACCGAACGUGACUAGGGUGCAAUUA ...................................(((.(((.(((((((...-)).....((((.......))))))).)).))))))..... ( -16.60) >DroSec_CAF1 19538 93 - 1 UUUAAUUAUAAUCCAUAAUCCAUAUAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAAUCGAACGUGACUAGGUUGCAAUUA ...................................((((((..(((((((...-)).....((((.......))))))).)).))))))..... ( -16.20) >DroSim_CAF1 19759 93 - 1 UUUAAUUAUAAUCCAUAAUCCAUAUAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCCAAAUCGAACGUGACUAGGUUGCAAUUA ...................................((((((..(((((((...-)).....((((.......))))))).)).))))))..... ( -16.20) >DroEre_CAF1 22914 80 - 1 CACUAUUG----CCAUAAU---------CCACAAGGCAACCUAAGCACGCCCA-GCGUUAGGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUA .....(((----((.....---------......))))).....((((.((..-..((((.((((.......)))).))))..))))))..... ( -20.30) >DroYak_CAF1 20888 81 - 1 UUCAAUUG----CCAUAAU---------CCACAAGGCAACCUAGGCACGCCCAGGCGUAAAGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUA ...(((((----((.....---------((....))...(((((.(((((....)).....((((.......))))))).)))))).)))))). ( -22.20) >consensus UUUAAUUAUAAUCCAUAAUCCAU_UA__CCACAAGGCAACCCAAGCACGCCCA_GCGUAAAGUUCCCAAAUCGAACGUGACUAGGGUGCAAUUA ...................................(((.(((.(((((((....)).....((((.......))))))).)).))))))..... (-15.04 = -15.04 + 0.00)

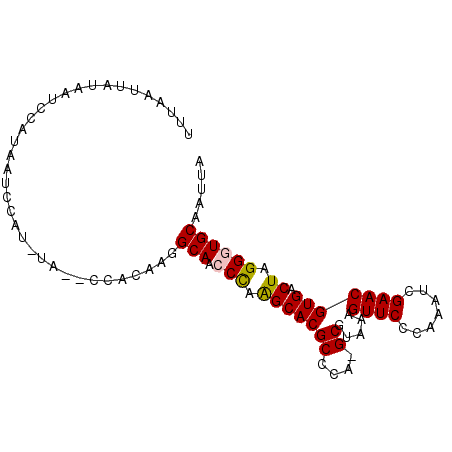

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:22 2006