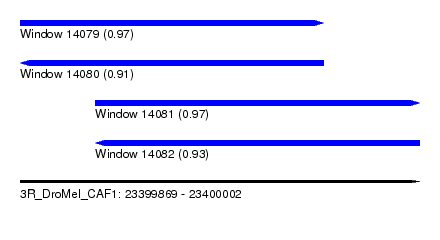
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,399,869 – 23,400,002 |
| Length | 133 |
| Max. P | 0.974457 |

| Location | 23,399,869 – 23,399,970 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -27.46 |
| Energy contribution | -28.50 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23399869 101 + 27905053 --------GGAGGAGUUG-----CUUCUUC-GCCAGAAGACAAAGUUGGCAACUCGAUUGAAAAAGUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG --------....((((((-----(((((((-....))))).......))))))))(((.(......(((((.((((.......)))).)))))....(((....)))..).))). ( -31.01) >DroSec_CAF1 14417 103 + 1 --------GAAGGAGUAGUUCUUCUUCUUG-GCCAGAAGACAAAGUUGGCAACUCGAUGGCA---GUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG --------(((((((.....)))))))(((-(((((....((..(((((((((((....).)---)))))).((((.......)))).....)))..)).))))))))....... ( -35.70) >DroSim_CAF1 14413 106 + 1 --------GAAGGAGUAGUUCUUCUUCUUG-GCCAGAAGACAAAGUUGGCAACUCGAUGGCAAAAGUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG --------(((((((.....)))))))(((-(((((....((...(((.((......)).)))...(((((.((((.......)))).)))))....)).))))))))....... ( -35.70) >DroEre_CAF1 17516 105 + 1 -------AGUAGGAGAA--UCUUGCGCUUG-GCCAGAAGACAAAGUUGGCAACUCGAUUGAAAAAGUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG -------.(((((....--.)))))(((((-(((((...........(....).(((((.....)))))))))))))))....((((.(((........))).))))........ ( -34.10) >DroYak_CAF1 15371 113 + 1 AGCGGAAAGUAGGAGCA--UCUUGCGCUUGGGCCAGAAGACAAAGUUGGCAACUCGAUUGAAAAAGUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG ...((..(((....((.--....))(((...((((...(((...((..((((((..........))))))..))...)))...)))).)))........)))..))......... ( -32.40) >consensus ________GAAGGAGUAG_UCUUCUUCUUG_GCCAGAAGACAAAGUUGGCAACUCGAUUGAAAAAGUUGCUGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCG ........(((((((.....)))))))....((((...(((...((..((((((..........))))))..))...)))...)))).(((...(....)...)))......... (-27.46 = -28.50 + 1.04)

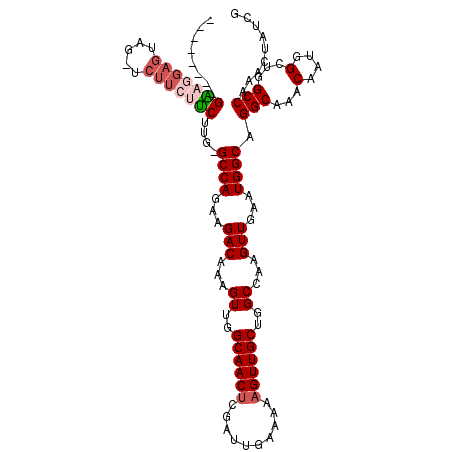
| Location | 23,399,869 – 23,399,970 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23399869 101 - 27905053 CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAACUUUUUCAAUCGAGUUGCCAACUUUGUCUUCUGGC-GAAGAAG-----CAACUCCUCC-------- .((.(((((((((((...(((..((.....))..))).)))))))..))))...))....(((((((...(((((((....)))-))))..)-----))))))....-------- ( -32.00) >DroSec_CAF1 14417 103 - 1 CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAAC---UGCCAUCGAGUUGCCAACUUUGUCUUCUGGC-CAAGAAGAAGAACUACUCCUUC-------- .(((((((((((.((.((.....)).))))).(((..(((((((((((((---(.......))))))((....)).....))))-))))..))))))))).))....-------- ( -31.10) >DroSim_CAF1 14413 106 - 1 CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAACUUUUGCCAUCGAGUUGCCAACUUUGUCUUCUGGC-CAAGAAGAAGAACUACUCCUUC-------- .(((((((((((.((.((.....)).))))).(((..(((((((((((((((........)))))))((....)).....))))-))))..))))))))).))....-------- ( -32.10) >DroEre_CAF1 17516 105 - 1 CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAACUUUUUCAAUCGAGUUGCCAACUUUGUCUUCUGGC-CAAGCGCAAGA--UUCUCCUACU------- .....((((((((((.((..(((.....((((.......))))..(((((((........))))))).)))..))....)))))-)))))......--..........------- ( -31.60) >DroYak_CAF1 15371 113 - 1 CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAACUUUUUCAAUCGAGUUGCCAACUUUGUCUUCUGGCCCAAGCGCAAGA--UGCUCCUACUUUCCGCU .....((((((((((.((..(((.....((((.......))))..(((((((........))))))).)))..))....)))).))))))((....--.)).............. ( -28.90) >consensus CGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCAGCAACUUUUUCAAUCGAGUUGCCAACUUUGUCUUCUGGC_CAAGAAGAAGA_CUACUCCUAC________ .....(..((((.((.((.....)).))))))..)..(((((((((((((((........)))))))((....)).....)))).)))).......................... (-21.64 = -22.24 + 0.60)

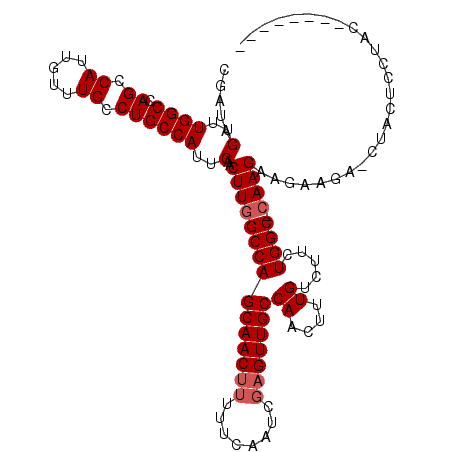
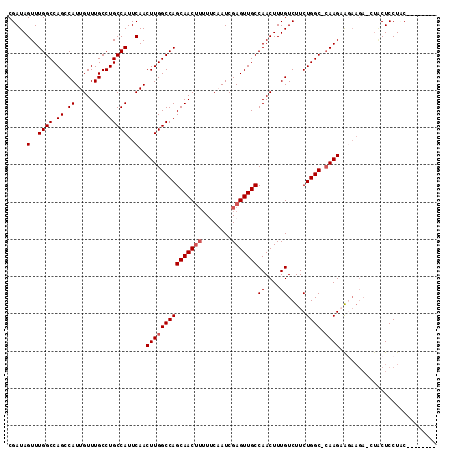
| Location | 23,399,894 – 23,400,002 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -19.43 |
| Energy contribution | -21.42 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23399894 108 + 27905053 ACAAAGUUGGCAACUCGAUUGAAAAAGUUGC---UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUGGCAAUAAAAA---GAG .....((((.(((((((((.((.....((((---(.((((.......)))).))))).((((.(.(((.....))).).))))...)).))))))))).)))).....---... ( -33.70) >DroSec_CAF1 14447 105 + 1 ACAAAGUUGGCAACUCGAUGGCA---GUUGC---UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUGGCAAUAAAAA---GAA .....((((.((((((((..(((---.((((---(.((((.......)))).))))).((((.(.(((.....))).).)))).)).)..)))))))).)))).....---... ( -36.10) >DroSim_CAF1 14443 108 + 1 ACAAAGUUGGCAACUCGAUGGCAAAAGUUGC---UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUGGCAAUAAAAA---AAA .....((((.((((((((((((...((((((---(.((((.......)))).)))).......))).)))((((.(((....)))))))))))))))).)))).....---... ( -36.41) >DroEre_CAF1 17545 111 + 1 ACAAAGUUGGCAACUCGAUUGAAAAAGUUGC---UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUUGCAAUAAAAACCAGAA .((((.((((((((((((((((.....((((---(.((((.......)))).)))))....(((....))).....)))))))).).))))))).))))............... ( -32.80) >DroYak_CAF1 15408 102 + 1 ACAAAGUUGGCAACUCGAUUGAAAAAGUUGC---UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUAGC---------UAGCA .....((((((((((((((.((.....((((---(.((((.......)))).))))).((((.(.(((.....))).).))))...)).)))))))).))---------)))). ( -34.40) >DroPer_CAF1 15927 90 + 1 AAAAAGUUGGCAGCUCGUUGGAAAUUGUUGAAGUUUUCACAGUUAAAUGGCA--------------GGCCAAACUAUCGAUUGGUGUCC-------UGCCAAUCCAAG---AAA ......((((.((((.((.(((((((.....)))))))))))))...(((((--------------((....((((.....))))..))-------)))))..)))).---... ( -24.70) >consensus ACAAAGUUGGCAACUCGAUGGAAAAAGUUGC___UGGCCAAGUUGAAUGGCAGGCAAACAAUGGCUGGCCAAACUAUCGAUUGAUGUUUGUCGAGUUGGCAAUAAAAA___GAA .....((((.(((((((((.....................((((...((((.(((........))).))))))))(((....)))....))))))))).))))........... (-19.43 = -21.42 + 1.99)

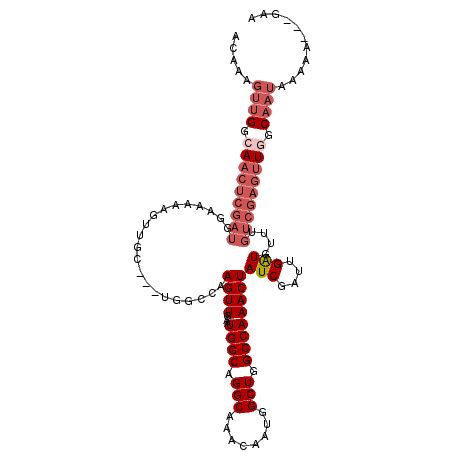
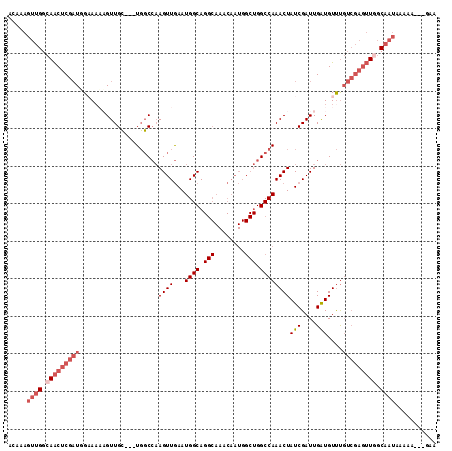
| Location | 23,399,894 – 23,400,002 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -15.27 |
| Energy contribution | -16.33 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23399894 108 - 27905053 CUC---UUUUUAUUGCCAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA---GCAACUUUUUCAAUCGAGUUGCCAACUUUGU ...---......(((.((((((((((((((((....))).)))))............((((..((((.......))))..---)))).........)))))))).)))...... ( -28.10) >DroSec_CAF1 14447 105 - 1 UUC---UUUUUAUUGCCAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA---GCAAC---UGCCAUCGAGUUGCCAACUUUGU ...---......(((.((((((((((((((((....))).)))))(.(((.......((((..((((.......))))..---)))))---)).).)))))))).)))...... ( -28.31) >DroSim_CAF1 14443 108 - 1 UUU---UUUUUAUUGCCAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA---GCAACUUUUGCCAUCGAGUUGCCAACUUUGU ...---......(((.((((((((((((((((....))).)))))....((.(..(((.((..((((.......))))..---)))))..).))..)))))))).)))...... ( -29.50) >DroEre_CAF1 17545 111 - 1 UUCUGGUUUUUAUUGCAAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA---GCAACUUUUUCAAUCGAGUUGCCAACUUUGU ..............((((.(((((((((((((....))).)))))............((((..((((.......))))..---)))).........)))))))))......... ( -25.60) >DroYak_CAF1 15408 102 - 1 UGCUA---------GCUAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA---GCAACUUUUUCAAUCGAGUUGCCAACUUUGU .....---------((.(((((((((((((((....))).)))))............((((..((((.......))))..---)))).........)))))))))......... ( -25.80) >DroPer_CAF1 15927 90 - 1 UUU---CUUGGAUUGGCA-------GGACACCAAUCGAUAGUUUGGCC--------------UGCCAUUUAACUGUGAAAACUUCAACAAUUUCCAACGAGCUGCCAACUUUUU .((---((((((.(((((-------((...((((.(....).))))))--------------))))).......((....))..........))))).)))............. ( -21.90) >consensus UUC___UUUUUAUUGCCAACUCGACAAACAUCAAUCGAUAGUUUGGCCAGCCAUUGUUUGCCUGCCAUUCAACUUGGCCA___GCAACUUUUUCAAUCGAGUUGCCAACUUUGU ............(((.((((((((................(..((((..((........))..))))..)......((.....))...........)))))))).)))...... (-15.27 = -16.33 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:16 2006