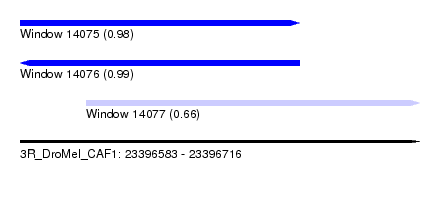
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,396,583 – 23,396,716 |
| Length | 133 |
| Max. P | 0.989288 |

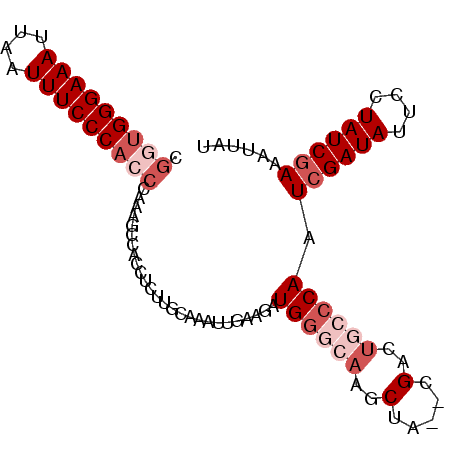
| Location | 23,396,583 – 23,396,676 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -16.56 |
| Energy contribution | -18.76 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23396583 93 + 27905053 CGGUGGGAAAUUAAUUUCCCACGAUAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCUA--CGACUGCCCAAUCGAUAUUACUAUCGAAAUCAU ..((((((((....))))))))(((........((((........))))((((((.....--....)))))).((((((....)))))).))).. ( -27.00) >DroSec_CAF1 11198 93 + 1 CGCUGGGAAAUUAAUUUCCCACCCAAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCUA--CGAUUGCCCAAUCGAUAUUCCUAUCGAAAUUAU ...(((((((....)))))))............((((........))))(((((((....--...))))))).((((((....))))))...... ( -24.20) >DroSim_CAF1 11219 93 + 1 CGCUGGGAAAUUAAUUUCCCACCCAAAGCCACCUCUUGCAAAGUGAAGAUGGGCAAGCUA--CGACUGCCCAAUCGAUAUUUCUAUCAAAAUUAU ((((((((((....)))))).......((........))..))))..((((((((.....--....))))).)))((((....))))........ ( -22.10) >DroEre_CAF1 14291 92 + 1 UGGGGGGAAAUUAAUUUCCCACCCAAAACCACC-CUCCCCAAUGGGAGGUGGCAAGGCUA--CGACUGCACAAUCGAUAUUCCUAUCGAACUUAU ((((((((((....)))))).))))...(((((-.((((....))))))))).(((((..--.....))....((((((....)))))).))).. ( -35.10) >DroYak_CAF1 12127 94 + 1 UGGUGGGAAAUUAAUUUCCCACCCAACGCCACC-CGCCCAAAAUGGAAGUGGCAUAGCUAGAUGACUGCCCAAUCGAUAUUGCUAUCGAACUUAU .(((((((((....)))))))))....(((((.-..((......))..)))))...((.((....))))....((((((....))))))...... ( -29.60) >consensus CGGUGGGAAAUUAAUUUCCCACCCAAAGCCACCUCUUGCAAAUUGAAGAUGGGCAAGCUA__CGACUGCCCAAUCGAUAUUCCUAUCGAAAUUAU .(((((((((....)))))))))..........................((((((..(.....)..)))))).((((((....))))))...... (-16.56 = -18.76 + 2.20)

| Location | 23,396,583 – 23,396,676 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -21.72 |
| Energy contribution | -23.24 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23396583 93 - 27905053 AUGAUUUCGAUAGUAAUAUCGAUUGGGCAGUCG--UAGCUUGCCCAUCUUCAAUUUGCAAGAGGCGGCUUAUCGUGGGAAAUUAAUUUCCCACCG ......((((((....))))))..((..(((((--(..(((((.............)))))..))))))..))((((((((....)))))))).. ( -32.32) >DroSec_CAF1 11198 93 - 1 AUAAUUUCGAUAGGAAUAUCGAUUGGGCAAUCG--UAGCUUGCCCAUCUUCAAUUUGCAAGAGGCGGCUUUGGGUGGGAAAUUAAUUUCCCAGCG ......((((((....)))))).(((((((...--....)))))))....(((.((((.....))))..)))(.(((((((....))))))).). ( -28.20) >DroSim_CAF1 11219 93 - 1 AUAAUUUUGAUAGAAAUAUCGAUUGGGCAGUCG--UAGCUUGCCCAUCUUCACUUUGCAAGAGGUGGCUUUGGGUGGGAAAUUAAUUUCCCAGCG ......((((((....)))))).((((((((..--..)).))))))...(((((((....))))))).....(.(((((((....))))))).). ( -30.70) >DroEre_CAF1 14291 92 - 1 AUAAGUUCGAUAGGAAUAUCGAUUGUGCAGUCG--UAGCCUUGCCACCUCCCAUUGGGGAG-GGUGGUUUUGGGUGGGAAAUUAAUUUCCCCCCA ...........(((.....(((((....)))))--...))).((((((((((....)))).-))))))..((((.((((((....)))))))))) ( -35.80) >DroYak_CAF1 12127 94 - 1 AUAAGUUCGAUAGCAAUAUCGAUUGGGCAGUCAUCUAGCUAUGCCACUUCCAUUUUGGGCG-GGUGGCGUUGGGUGGGAAAUUAAUUUCCCACCA ......((((((....))))))...(((((....)).)))(((((((((((......)).)-))))))))..(((((((((....))))))))). ( -37.30) >consensus AUAAUUUCGAUAGGAAUAUCGAUUGGGCAGUCG__UAGCUUGCCCAUCUUCAAUUUGCAAGAGGUGGCUUUGGGUGGGAAAUUAAUUUCCCACCG ......((((((....)))))).(((((((.(.....).)))))))..........................(((((((((....))))))))). (-21.72 = -23.24 + 1.52)

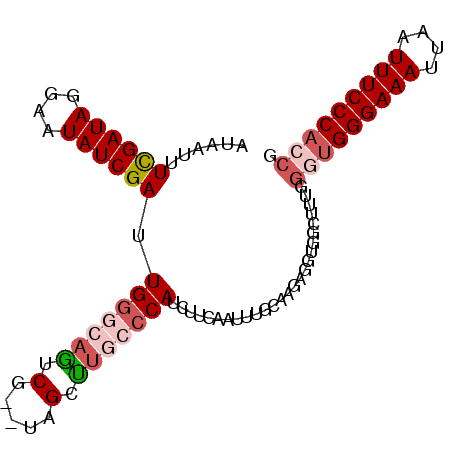
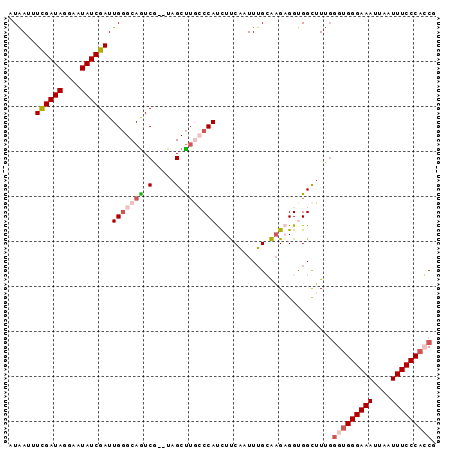
| Location | 23,396,605 – 23,396,716 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -13.66 |
| Energy contribution | -15.14 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23396605 111 + 27905053 GAUAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCUA--CGACUGCCCAAUCGAUAUUACUAUCGAAAUCAUCGAUGCUUCUUUAUCGAUAAACUAAAAACACUUGAUUCCU (((((((.((.....))..(((((.((((((((.....--....)))))..((((((....)))))).))).))))))))....))))......................... ( -22.00) >DroSec_CAF1 11220 102 + 1 CCAAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCUA--CGAUUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGACACUUCUUUAUCGAUAAG---------CUUGAUUCCU ...........((..((..........(((((((....--...))))))).((((((....))))))..(((((((..........))))))))---------)..))..... ( -23.80) >DroSim_CAF1 11241 102 + 1 CCAAAGCCACCUCUUGCAAAGUGAAGAUGGGCAAGCUA--CGACUGCCCAAUCGAUAUUUCUAUCAAAAUUAUCGACGCUUCAUUAUCGAUAAG---------CUUGAUUCCU ...((((......(((...(((((((.((((((.....--....)))))).((((((.............))))))..)))))))..)))...)---------)))....... ( -21.32) >DroEre_CAF1 14313 110 + 1 CCAAAACCACC-CUCCCCAAUGGGAGGUGGCAAGGCUA--CGACUGCACAAUCGAUAUUCCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUGAACCUAACAUAAUUCAUUCCU ......(((((-.((((....)))))))))..(((...--...........((((((....))))))..(((((((((......))))))))).)))................ ( -28.70) >DroYak_CAF1 12149 112 + 1 CCAACGCCACC-CGCCCAAAAUGGAAGUGGCAUAGCUAGAUGACUGCCCAAUCGAUAUUGCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUAAACAUAAAAUACUUGAUUCUU .....(((((.-..((......))..)))))........(((.........((((((....))))))..(((((((((......))))))))).)))................ ( -23.40) >consensus CCAAAGCCACCUCUUGCAAAUUGAAGAUGGGCAAGCUA__CGACUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGAUGCUUCUUUAUCGAUAAAC__AA_A_ACUUGAUUCCU ...........................((((((..(.....)..)))))).((((((....))))))..(((((((((......))))))))).................... (-13.66 = -15.14 + 1.48)

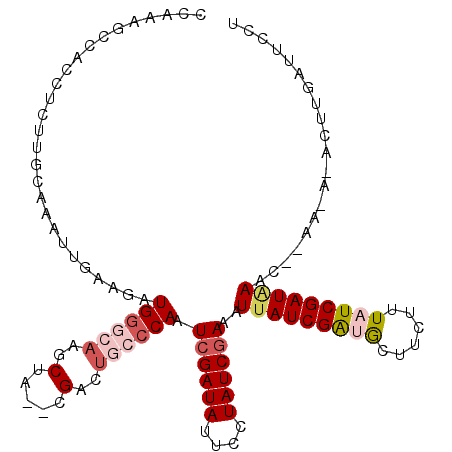
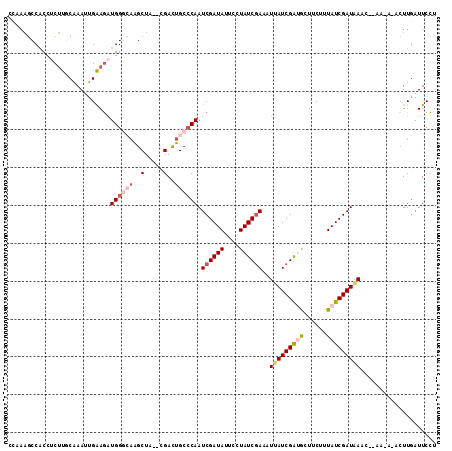
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:11 2006