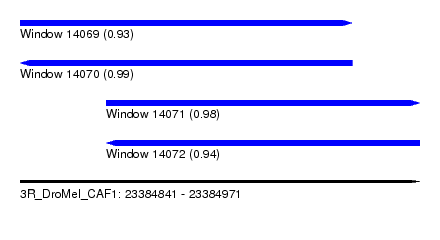
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,384,841 – 23,384,971 |
| Length | 130 |
| Max. P | 0.991888 |

| Location | 23,384,841 – 23,384,949 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23384841 108 + 27905053 GUUCAUCGUUCGACUCGGUGUGACCGUUCGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAAUUCGAUGUUUUCAACA (((......((((...(((((((((((((((((...............)))))))......((((.(((....)))))))))))))))))...)))).......))). ( -39.88) >DroSec_CAF1 10442 108 + 1 GUUCAUCGUUCGACUCGGUGUGACCGUUCGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCUUUAUGGCAGUGCGUUCACACCAUUUCUAUGUAUUCAACA ................(((((((.(((((((((...............)))))))......((((.(((....))))))))).))))))).................. ( -27.56) >DroSim_CAF1 10664 108 + 1 GUUCAUCGUUCGACUCGGUGUGACCGUUCGCAGGAUAGUGCCAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAUUUCUAUGUUUUCAACA ................(((((((((((((((((..((.......))..)))))))......((((.(((....))))))))))))))))).................. ( -38.80) >DroYak_CAF1 10594 106 + 1 GUUCAUCGUUCGACUCAGUGUGACCGUUCAGAGGGCAGUGU-GUUUAGCCGUGGAGUUUACCACUUGCG-UCUAACAGUGCGGUCACACUCAUUCUAUGUUUUCAGUA ...........((.(.(((((((((...(....)(((.(((-.....((.((((......))))..)).-....))).)))))))))))).).))............. ( -29.10) >consensus GUUCAUCGUUCGACUCGGUGUGACCGUUCGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAAUUCUAUGUUUUCAACA ................(((((((((((((((((...............)))))))......((((.(((....))))))))))))))))).................. (-29.58 = -29.96 + 0.38)

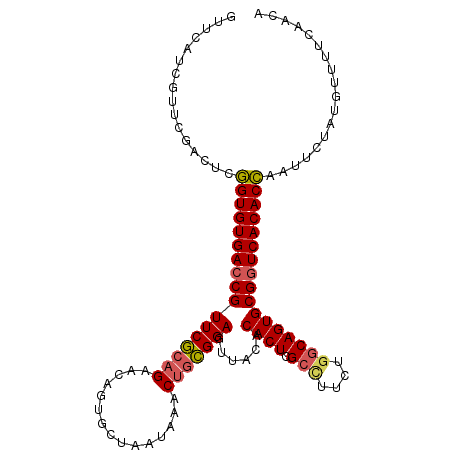

| Location | 23,384,841 – 23,384,949 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -30.17 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23384841 108 - 27905053 UGUUGAAAACAUCGAAUUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUUCUGCGAACGGUCACACCGAGUCGAACGAUGAAC ...........((((.(((((((((((((((((((....))).))))).......(((((..((.......))..)))))...))))))))))).))))......... ( -43.10) >DroSec_CAF1 10442 108 - 1 UGUUGAAUACAUAGAAAUGGUGUGAACGCACUGCCAUAAAGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUUCUGCGAACGGUCACACCGAGUCGAACGAUGAAC .........(((.((..(((((((((((((((((......)).))))))).....(((((..((.......))..))))).....))))))))..)).....)))... ( -31.90) >DroSim_CAF1 10664 108 - 1 UGUUGAAAACAUAGAAAUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUGGCACUAUCCUGCGAACGGUCACACCGAGUCGAACGAUGAAC .........(((.((..((((((((((((((((((....))).))))).......(((((..((.......))..)))))...))))))))))..)).....)))... ( -39.40) >DroYak_CAF1 10594 106 - 1 UACUGAAAACAUAGAAUGAGUGUGACCGCACUGUUAGA-CGCAAGUGGUAAACUCCACGGCUAAAC-ACACUGCCCUCUGAACGGUCACACUGAGUCGAACGAUGAAC .........(((.((.(.((((((((((.....(((((-.....((((......))))(((.....-.....))).))))).)))))))))).).)).....)))... ( -32.00) >consensus UGUUGAAAACAUAGAAAUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUCCUGCGAACGGUCACACCGAGUCGAACGAUGAAC .............((.((((((((((((((((((......)).))))........(((((..((.......))..)))))..)))))))))))).))........... (-30.17 = -30.43 + 0.25)

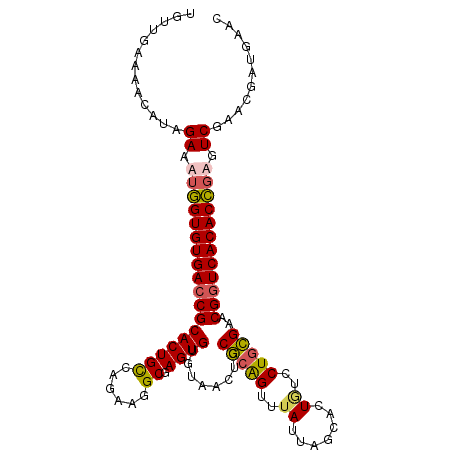
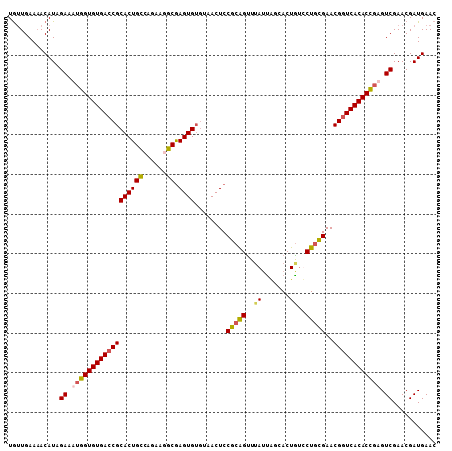
| Location | 23,384,869 – 23,384,971 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23384869 102 + 27905053 CGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAAUUCGAUGUUUUCAACAUUCAUAGAAAUGGUUGAUAACA (((((...............)))))..(((.(.((((.(((....))))))).)((((.((((.((((((((.....)))))....))).))))))))))). ( -26.36) >DroSec_CAF1 10470 102 + 1 CGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCUUUAUGGCAGUGCGUUCACACCAUUUCUAUGUAUUCAACAUUCAUAGAAAUGGUUCAUAGCU .((.((((.(..((...........((((.....))))(((....)))))..)))))..((((((((((((...........)))))))))))).....)). ( -31.00) >DroSim_CAF1 10692 102 + 1 CGCAGGAUAGUGCCAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAUUUCUAUGUUUUCAACAUUCAUAGAAAUGGUUCAUAGCU (((((..((.......))..))))).((((((.((((.(((....))))))).).....((((((((((((...........))))))))))))...))))) ( -34.10) >DroYak_CAF1 10622 100 + 1 CAGAGGGCAGUGU-GUUUAGCCGUGGAGUUUACCACUUGCG-UCUAACAGUGCGGUCACACUCAUUCUAUGUUUUCAGUAUUCAUAGAAAUGGUUGAUGGCU ......(((.(((-.....((.((((......))))..)).-....))).)))(((((((..(((((((((..(.....)..))))).))))..)).))))) ( -28.30) >consensus CGCAGAACAGUGCUAAUAAACUGCGGAGUUACACACUCGCCUUCUGGCAGUGCGGUCACACCAAUUCUAUGUUUUCAACAUUCAUAGAAAUGGUUCAUAGCU .........(((.......((((((((((.....))))(((....)))..)))))))))((((((((((((...........))))))))))))........ (-21.95 = -22.63 + 0.69)

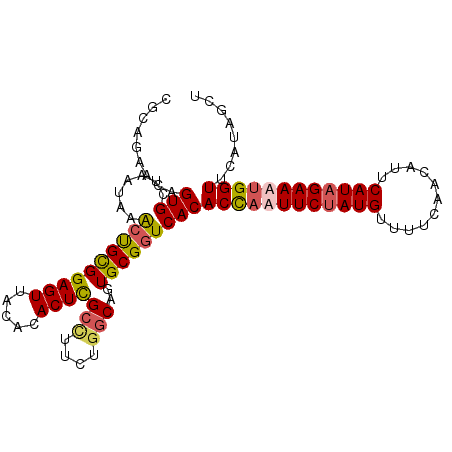
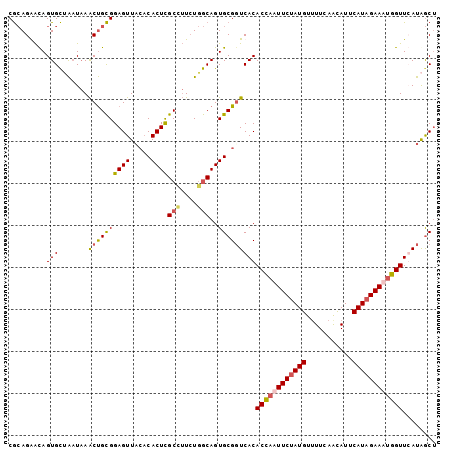
| Location | 23,384,869 – 23,384,971 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -21.65 |
| Energy contribution | -23.02 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23384869 102 - 27905053 UGUUAUCAACCAUUUCUAUGAAUGUUGAAAACAUCGAAUUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUUCUGCG ((((.(((((.((((....))))))))).)))).......((.((...(((((((((....))).))))))..)).))((((..((.......))..)))). ( -29.00) >DroSec_CAF1 10470 102 - 1 AGCUAUGAACCAUUUCUAUGAAUGUUGAAUACAUAGAAAUGGUGUGAACGCACUGCCAUAAAGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUUCUGCG .(((((((((((((((((((.((.....)).))))))))))(((.(((((((((((......)).)))))))...))))).))))).))))........... ( -32.30) >DroSim_CAF1 10692 102 - 1 AGCUAUGAACCAUUUCUAUGAAUGUUGAAAACAUAGAAAUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUGGCACUAUCCUGCG ........((((((((((((...........))))))))))))((...(((((((((....))).))))))..))..(((((..((.......))..))))) ( -33.40) >DroYak_CAF1 10622 100 - 1 AGCCAUCAACCAUUUCUAUGAAUACUGAAAACAUAGAAUGAGUGUGACCGCACUGUUAGA-CGCAAGUGGUAAACUCCACGGCUAAAC-ACACUGCCCUCUG ((((.........(((((((...........))))))).((((...(((((..(((....-.))).)))))..))))...))))....-............. ( -22.60) >consensus AGCUAUCAACCAUUUCUAUGAAUGUUGAAAACAUAGAAAUGGUGUGACCGCACUGCCAGAAGGCGAGUGUGUAACUCCGCAGUUUAUUAGCACUGUCCUGCG ........((((((((((((...........))))))))))))((...((((((((......)).))))))..))..(((((..((.......))..))))) (-21.65 = -23.02 + 1.38)

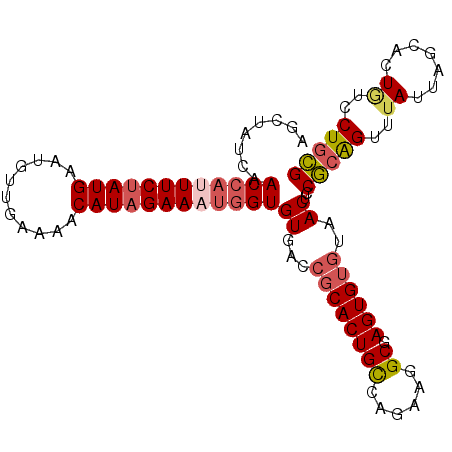
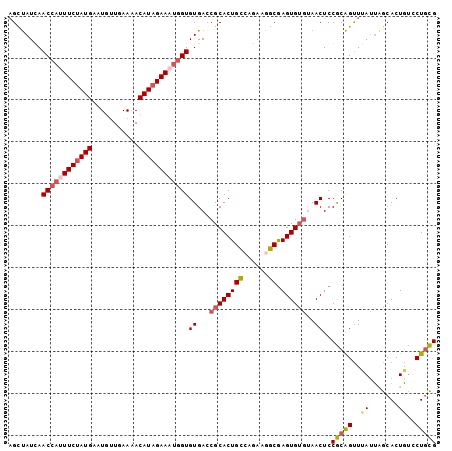
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:06 2006