
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,384,508 – 23,384,620 |
| Length | 112 |
| Max. P | 0.725181 |

| Location | 23,384,508 – 23,384,620 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23384508 112 - 27905053 AAUU---AAUGUCGAAUUGCGCACAAUUUAAUCUUAAUUUUAUUAGUGGUAGUAUGUGCAGUUGUUUGUGGAGCCGCGAAAAGUUCG-CAAAGUUCGCUGAUUUUGCGAACGGGAG- ((((---((.((..(((((....)))))..)).))))))......(((((..((((.((....)).))))..))))).....(((((-(((((((....)))))))))))).....- ( -28.80) >DroPse_CAF1 10159 103 - 1 AAUC---GAAAUUAUAUUGCGCACAAU-CAUCAUU---AUUAGUACGAGUAUUAUGUGCAGUUGUCUGCGGAGGCGCGCAAAGUACGGCCAAG-------AAGUCGUGGACAGCGGA ....---..........((((((.(((-..((...---........))..))).))))))((((((..(((.(((.((.......)))))...-------...)))..))))))... ( -26.90) >DroSec_CAF1 10109 112 - 1 AAUU---AAUGUCGAAUUGCGCACAAUUUAAUUUUAAUUUCACUAGUGGUAGUAUGUGCAGUUGUUUGUGGCGCCGCGAAAAGUUCG-CAAAGUUCGCUGAUUUUGCGAACGGCAG- ....---..((((.(((((((((.((((.......))))..((((....)))).))))))))).((((((....))))))..(((((-(((((((....)))))))))))))))).- ( -32.90) >DroSim_CAF1 10331 112 - 1 AAUU---AAUGUCGAAUUGCGCACAAUUUAAUUUUAAUUUCACUAGUGGUAGUAUGUGCAGUUGUUUGUGGCGCCGCGAAAAGUUCG-CAAAGUUCGCUGAUUUUGCGAACGGCAG- ....---..((((.(((((((((.((((.......))))..((((....)))).))))))))).((((((....))))))..(((((-(((((((....)))))))))))))))).- ( -32.90) >DroYak_CAF1 10260 111 - 1 AAUU---AAUGUCGAAUUGCGCACAAA-UAUUAUUCAUUUCACUAGUGGUAGUAUGUGCAGUUGUUUGUGGCGACGCGAAAAGUUCG-CAAAGUUCGCUGAUUUUGCGAACAGGAG- ....---.......(((((((((....-((((((.((((.....))))))))))))))))))).((((((....))))))..(((((-(((((((....)))))))))))).....- ( -34.50) >DroAna_CAF1 8410 105 - 1 AAUCGAAAAAGUGGAAUUGCGCACAAU-UAUUCGCAGCAGCACUGCUGCUAGUAUGUGCAGUUGUUUGUGGCGGCGCGCAAAGUUCG-CUAAGUUCG---------CGGACAGCAG- ....(((..((((((.(((((((((..-((((.((((((....)))))).))))))))).(((((.....)))))..))))..))))-))...)))(---------(.....))..- ( -36.90) >consensus AAUU___AAUGUCGAAUUGCGCACAAU_UAAUAUUAAUUUCACUAGUGGUAGUAUGUGCAGUUGUUUGUGGCGCCGCGAAAAGUUCG_CAAAGUUCGCUGAUUUUGCGAACAGCAG_ .........((.((((((..(((((................((((....)))).))))).....((((((....)))))).)))))).))..((((((.......))))))...... (-17.50 = -18.00 + 0.49)

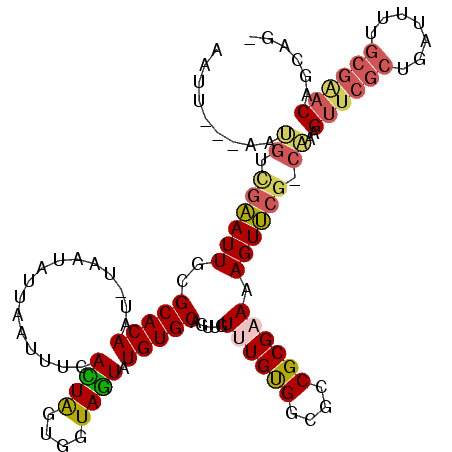
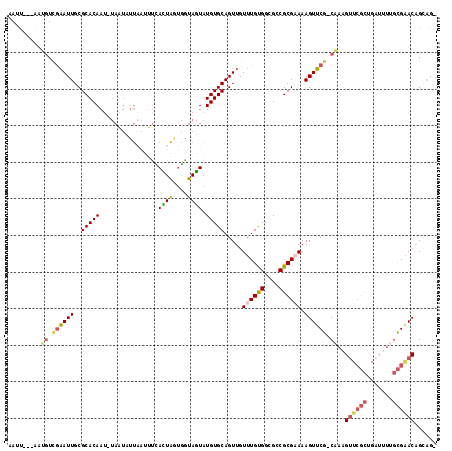
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:03 2006