
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,376,290 – 23,376,402 |
| Length | 112 |
| Max. P | 0.527865 |

| Location | 23,376,290 – 23,376,402 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.25 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
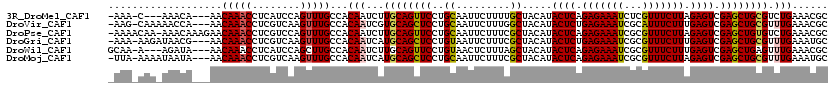
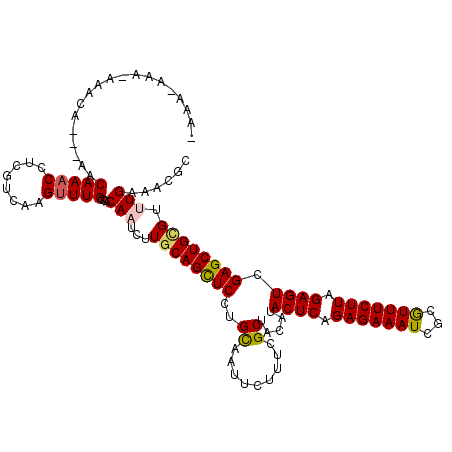
>3R_DroMel_CAF1 23376290 112 + 27905053 -AAA-C---AAACA---AACAAACCUCAUCCAGUUUGCCACAAUCUUGCAGUUCCUGCAAUUCUUUUGCUACAUACUCAGAGAAAUCUCGUUUCUUAGAGUCGAGCUGCGUCUGAAACGC -...-(---(.((.---..(((((........)))))..........(((((((..((((.....)))).....((((.(((((((...))))))).)))).))))))))).))...... ( -26.60) >DroVir_CAF1 98617 115 + 1 -AAG-CAAAAACCA---AACAAACCUCGUCAAGUUUGCCACAAUCGUGCAGCUCCUGCAAUUCUUUGGCUACAUACUCUGAGAAAUCGCAUUUCUUUGAGUCGAGCUGCGUUUGAAACGC -...-.......((---(((....((((....((..((((......(((((...)))))......)))).))..((((.(((((((...))))))).))))))))....)))))...... ( -28.00) >DroPse_CAF1 1949 118 + 1 -AAAACAA-AAACAAAGAACAAACCUCGUCCAGUUUGCCACAAUCUUGCAGUUCCUGCAAUUCUUUCGCUACAUACUCAGAGAAAUCGCGUUUCUUAGAGUCGAGCUGUGUCUGAAACGC -.......-......(((.(((((........))))).((((...((((((...)))))).......(((....((((.(((((((...))))))).))))..))))))))))....... ( -23.60) >DroGri_CAF1 36184 115 + 1 -AAA-AAGAUAACG---AACAAACCUCGUCAAGUUUGCCACAAUCAUGCAGCUCCUGUAAUUCUUUCGCUACAUACUCUGAGAAAUCGCGUUUCUUUGAGUCGAGCUGCGUUUGAAAUGC -...-.......((---(((((((........))))...........(((((((.((((..........)))).((((.(((((((...))))))).)))).))))))))))))...... ( -26.40) >DroWil_CAF1 1942 113 + 1 GCAA-A---AGAUA---AACAAACCUCAUCCAGCUUGCCACAAUCUUGCAGUUCCUGUAACUCUUUAGCUACAUACUCAGAGAAAUCGCGUUUCUUUGAGUCGAGCUGAGUUUGAAACGC ....-.---.....---..(((((.....................((((((...))))))....((((((....((((((((((((...))))))))))))..)))))))))))...... ( -26.40) >DroMoj_CAF1 40734 115 + 1 -UUA-AAAAUAAUA---AACAAACCUCGUCAAGUUUGCCACAAUCAUGCAGCUCCUGCAAUUCUUUCGCUACAUACUCAGAGAAAUCGCGUUUCUUAGAGUCGAGCUGCGUUUGAAAUGC -...-.........---..(((((........)))))...(((..(((((((((..((.........)).....((((.(((((((...))))))).)))).))))))))))))...... ( -25.90) >consensus _AAA_AAA_AAACA___AACAAACCUCGUCAAGUUUGCCACAAUCUUGCAGCUCCUGCAAUUCUUUCGCUACAUACUCAGAGAAAUCGCGUUUCUUAGAGUCGAGCUGCGUUUGAAACGC ...................(((((........)))))...(((...((((((((..((.........)).....((((.(((((((...))))))).)))).)))))))).)))...... (-21.30 = -21.25 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:59 2006