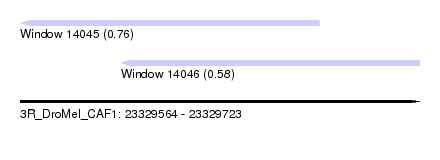
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,329,564 – 23,329,723 |
| Length | 159 |
| Max. P | 0.761228 |

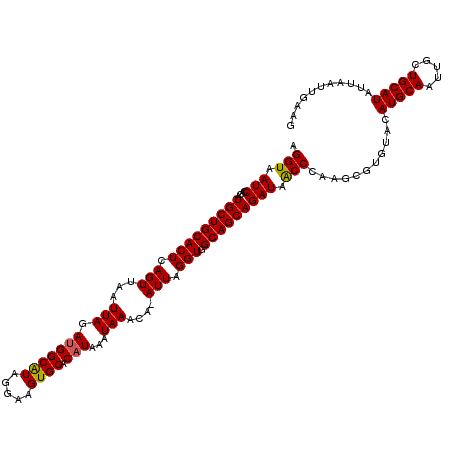
| Location | 23,329,564 – 23,329,683 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23329564 119 - 27905053 AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUAACCGACGCGUAUACAUGCAAUUGCUGCAUAUUAAUUGAAG .(((.(((....((((((((..((((..(((.(((((((.....)))).)))...)))..)-)))..)).))))))))).)))...........(((((.....)))))........... ( -28.40) >DroSec_CAF1 126844 119 - 1 AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCGUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUAACCCAAGCGUGUACAUGCAAUUGCUGCAUAUUAAUUGAAG .((....))..(((.(((((((.((((......))))....).))))))))).......((-(((((...(((((((......)..((((....))))...))))))...)))))))... ( -30.20) >DroSim_CAF1 131197 119 - 1 AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUAACCCAAGCGUGUACAUGCAAUUGCUGCAUAUUAAUUGAAG .((....))..(((.(((((((.((((......))))....).))))))))).......((-(((((...(((((((......)..((((....))))...))))))...)))))))... ( -29.10) >DroEre_CAF1 133212 119 - 1 AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUGGGUGGCAGCAGAUAGCCCAGUCGUGUACAUGCAAUUGCUGCAUAUUAAUUGAAG .(((.(((....((((((((((((((..(((.(((((((.....)))).)))...)))..)-))))))).))))))))).)))((((..(((....)))...)))).............. ( -33.00) >DroYak_CAF1 138810 120 - 1 AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGACGGCAUAGGAAGUGCACAUAAAUAAACAAAUUGGGUGGCAGCAGAUAACCCAGCCGUGUACAUGCAAUUGCUGCAUAUUAAUUGAAG .(((.(((....((((((((((((((..(((....((((.....)))).......)))...)))))))).))))))))).)))((((..(((....)))...)))).............. ( -30.00) >consensus AGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA_AUUAGGUGGCAGCAGAUAACCCAAGCGUGUACAUGCAAUUGCUGCAUAUUAAUUGAAG .(((.(((....(((((((((.(((...(((.(((((((.....)))).)))...)))....))).))).))))))))).)))...........(((((.....)))))........... (-24.60 = -24.48 + -0.12)


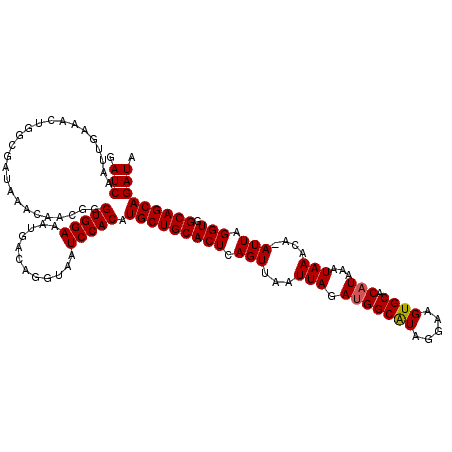
| Location | 23,329,604 – 23,329,723 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23329604 119 - 27905053 GAUCAAUUGAAACUGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUA .(((........(((...........)))(((((............))))).((((((((..((((..(((.(((((((.....)))).)))...)))..)-)))..)).))))))))). ( -27.20) >DroSec_CAF1 126884 119 - 1 GAUCAAUUGAAACUGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCGUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUA .(((........(((...........)))(((((............))))).((((((((..((((..(((.((((((.......))).)))...)))..)-)))..)).))))))))). ( -24.70) >DroSim_CAF1 131237 119 - 1 GAUCAAUUGAAACUGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUAGGUGGCAGCAGAUA .(((........(((...........)))(((((............))))).((((((((..((((..(((.(((((((.....)))).)))...)))..)-)))..)).))))))))). ( -27.20) >DroEre_CAF1 133252 119 - 1 GAUCAAUUGAAACAGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA-AUUGGGUGGCAGCAGAUA .(((.........................(((((............))))).((((((((((((((..(((.(((((((.....)))).)))...)))..)-))))))).))))))))). ( -31.50) >DroYak_CAF1 138850 120 - 1 GAUCAAUUGAAACAGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGACGGCAUAGGAAGUGCACAUAAAUAAACAAAUUGGGUGGCAGCAGAUA .(((.........................(((((............))))).((((((((((((((..(((....((((.....)))).......)))...)))))))).))))))))). ( -27.20) >consensus GAUCAAUUGAAACUGGCGAUAAACAACGGGUGGAAAUGACAGGUAAUCCACAUGCUGCACUCAGUUAAUUAGAUGGCAUAGGAAGUGCACAUAAAUAAACA_AUUAGGUGGCAGCAGAUA .(((.........................(((((............))))).(((((((((.(((...(((.(((((((.....)))).)))...)))....))).))).))))))))). (-23.20 = -23.24 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:42 2006