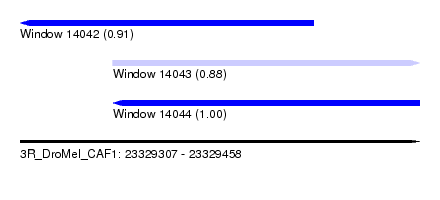
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,329,307 – 23,329,458 |
| Length | 151 |
| Max. P | 0.998464 |

| Location | 23,329,307 – 23,329,418 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23329307 111 - 27905053 AAAUGCUUUUCCACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCUCGAAGUGGAGGGCAAAAUGCAUCUGC---------CAACACCAACGCCAUUAUCCUGGGCAUAUGUCUAA ...............((((((((((((((((((......).))))....))).)))))))))).....((((.(((---------(.....................)))).)))).... ( -33.10) >DroSec_CAF1 126584 113 - 1 AAAUGCUUUUCCACUGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCGCAUCUGCCAA------CGCCAUUAUCCUGGGCAUAUGUCUAA ..((((........(((....(((((((.....)))))))..)))..(((.(((((((..((((.-((((.......)))).))))..------..))))).)).)))))))........ ( -35.90) >DroSim_CAF1 130931 119 - 1 AAAUGCUUUUGCACUGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCGCAUCUGCCAACGCCAACGCCAUUAUCCUGGGCAUAUGUCUAA ....((...((((.(((((((((((((.(((((......).)))).....)).))))))))))).-.))))...)).((((.((((...((....))((......)))))).)))).... ( -38.90) >DroEre_CAF1 132941 116 - 1 AAAUGCUUUUCUACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCAG---CGCCAACGCCAACGCCGUUAUCCUGGGCAUAUGUCUAA ...............((((((((((((.(((((......).)))).....)).))))))))))..-..((((.((((((---.(..(((((....).))))..))).)))).)))).... ( -34.90) >DroYak_CAF1 138544 116 - 1 AAAUGCUUUUCUACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCAA---CGCCAACGCCAACGCCAUUAUCCUGGGCAUAUGUCUAA ...............((((((((((((.(((((......).)))).....)).))))))))))..-..((((.((((..---.......((....))..........)))).)))).... ( -32.61) >consensus AAAUGCUUUUCCACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA_AUGCAUCUGCCAC___CGCCAACGCCAACGCCAUUAUCCUGGGCAUAUGUCUAA ..((((.........((((((((((((.(((((......).)))).....)).)))))))))).................................(((......)))))))........ (-29.30 = -29.14 + -0.16)

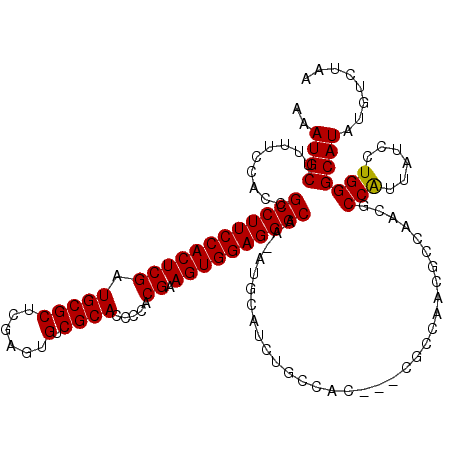
| Location | 23,329,342 – 23,329,458 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -33.90 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23329342 116 + 27905053 ----GCAGAUGCAUUUUGCCCUCCACUUCGAGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCGGUGGAAAAGCAUUUCAAAUGCAUUCACAAGCAGUAGCGUUUUAAAUUACUCCCA ----((...(((....((((.((((((.(((...((((.........)))).))))))))).))))(((((...((((.....)))).)))))..)))...))................. ( -37.90) >DroSec_CAF1 126618 119 + 1 UGCGGCAGAUGCAU-UUGCCCUCCACUUCGUGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCAGUGGAAAAGCAUUUCAAAUGCAUUCACAAGCAGGAGCGUUUUAAAUUACUCCCA (((....(((((((-(((((.(((((((.(((.(.(.((.....))).).))).))))))).)))....((((....))))))))))))).....)))((((............)))).. ( -38.60) >DroSim_CAF1 130971 119 + 1 UGCGGCAGAUGCAU-UUGCCCUCCACUUCGUGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCAGUGCAAAAGCAUUUCAAAUGCAUUCACAAGCAGGAGCGUUUUAAAUUACUCCCA (((....(((((((-(((((.(((((((.(((.(.(.((.....))).).))).))))))).)))(((((....)))))..))))))))).....)))((((............)))).. ( -41.30) >DroEre_CAF1 132979 118 + 1 -CUGGCAGAUGCAU-UUGCCCUCCACUUCGUGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCGGUAGAAAAGCAUUUCAAAUGCAUUCACGAGCAGGAGCGUUUUAAAUUACUCCCA -..((((((....)-)))))((((..((((((((.((((.(((((((.....)))))))((((((.........)).))))...))))))))))))..)))).................. ( -38.60) >DroYak_CAF1 138582 118 + 1 -UUGGCAGAUGCAU-UUGCCCUCCACUUCGUGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCGGUAGAAAAGCAUUUCAAAUGCAUUCACGAGCAGGAGCGUUUUAAAUUACUCCCA -..((((((....)-)))))((((..((((((((.((((.(((((((.....)))))))((((((.........)).))))...))))))))))))..)))).................. ( -38.60) >consensus _GCGGCAGAUGCAU_UUGCCCUCCACUUCGUGGGGUGCGACACUCGAGCGCAUCGAGUGGAAGGCGGUGGAAAAGCAUUUCAAAUGCAUUCACAAGCAGGAGCGUUUUAAAUUACUCCCA ....((...(((....((((.(((((((.....((((((.(......)))))))))))))).))))(((((...((((.....)))).)))))..)))...))................. (-33.90 = -34.26 + 0.36)

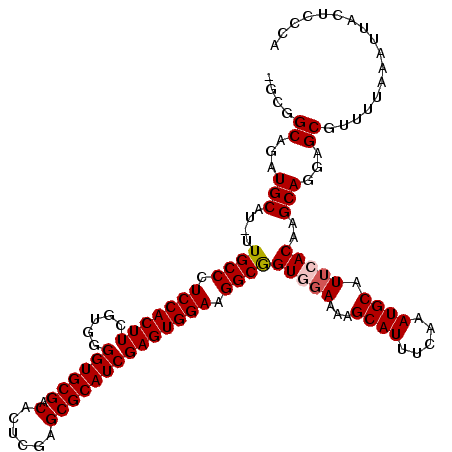
| Location | 23,329,342 – 23,329,458 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -41.86 |
| Consensus MFE | -36.08 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
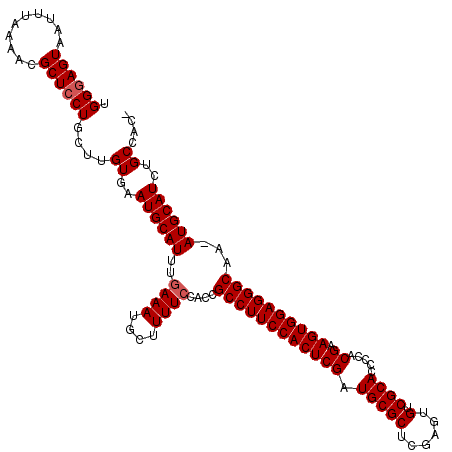
>3R_DroMel_CAF1 23329342 116 - 27905053 UGGGAGUAAUUUAAAACGCUACUGCUUGUGAAUGCAUUUGAAAUGCUUUUCCACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCUCGAAGUGGAGGGCAAAAUGCAUCUGC---- .((.((((...........)))).)).((..((((((((((((....))))....((((((((((((((((((......).))))....))).)))))))))).))))))))..))---- ( -38.50) >DroSec_CAF1 126618 119 - 1 UGGGAGUAAUUUAAAACGCUCCUGCUUGUGAAUGCAUUUGAAAUGCUUUUCCACUGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCGCA .((((((..........))))))((..((..((((((((((((....))))....((((((((((((.(((((......).)))).....)).))))))))))))-))))))..)).)). ( -42.80) >DroSim_CAF1 130971 119 - 1 UGGGAGUAAUUUAAAACGCUCCUGCUUGUGAAUGCAUUUGAAAUGCUUUUGCACUGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCGCA .((((((..........))))))((..((..((((((((....(((....)))..((((((((((((.(((((......).)))).....)).))))))))))))-))))))..)).)). ( -44.50) >DroEre_CAF1 132979 118 - 1 UGGGAGUAAUUUAAAACGCUCCUGCUCGUGAAUGCAUUUGAAAUGCUUUUCUACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCAG- .((((((..........)))))).((.((..((((((((((((....))))....((((((((((((.(((((......).)))).....)).))))))))))))-))))))..)).))- ( -41.80) >DroYak_CAF1 138582 118 - 1 UGGGAGUAAUUUAAAACGCUCCUGCUCGUGAAUGCAUUUGAAAUGCUUUUCUACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA-AUGCAUCUGCCAA- .((((((..........))))))....((..((((((((((((....))))....((((((((((((.(((((......).)))).....)).))))))))))))-))))))..))...- ( -41.70) >consensus UGGGAGUAAUUUAAAACGCUCCUGCUUGUGAAUGCAUUUGAAAUGCUUUUCCACCGCCUUCCACUCGAUGCGCUCGAGUGUCGCACCCCACGAAGUGGAGGGCAA_AUGCAUCUGCCAC_ .((((((..........))))))....((..((((((..((((....))))....((((((((((((.(((((......).)))).....)).))))))))))...))))))..)).... (-36.08 = -36.48 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:40 2006