
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,325,882 – 23,326,012 |
| Length | 130 |
| Max. P | 0.857650 |

| Location | 23,325,882 – 23,325,973 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23325882 91 - 27905053 UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGU--------AUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAU------------------GUU ((((((((........))))).))).((((((.((((((.....))))))))))--------))..............(((((.........))))------------------).. ( -24.70) >DroSec_CAF1 123188 91 - 1 UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGU--------AUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAU------------------GUU ((((((((........))))).))).((((((.((((((.....))))))))))--------))..............(((((.........))))------------------).. ( -24.70) >DroSim_CAF1 127549 91 - 1 UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGU--------AUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAU------------------GUU ((((((((........))))).))).((((((.((((((.....))))))))))--------))..............(((((.........))))------------------).. ( -24.70) >DroEre_CAF1 129518 99 - 1 UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGUAUGCCAGUAUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAU------------------GUU ((((((((........))))).))).((((((.((((((.....))))))))))))....(((((....)))))....(((((.........))))------------------).. ( -25.80) >DroYak_CAF1 134947 117 - 1 UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGUGUGCCUAUAUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAUGUUCUUCAUGUUGUUGAUGUU ((((((((........))))).))).((((((.((((((.....))))))))))))((.((((....)))).)).........(((((((.((.(((.....))).)).))))))). ( -29.10) >consensus UGUUGGCUAAAAUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGU________AUGCCUAUAUGCCUAGUAUCCGCGUUAAAAGGAU__________________GUU ((((((((........))))).)))...((((.((((((.....))))))))))...........(((..((((.........))))....)))....................... (-21.58 = -21.58 + 0.00)

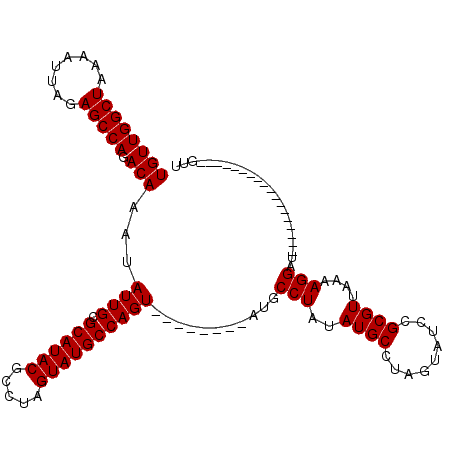
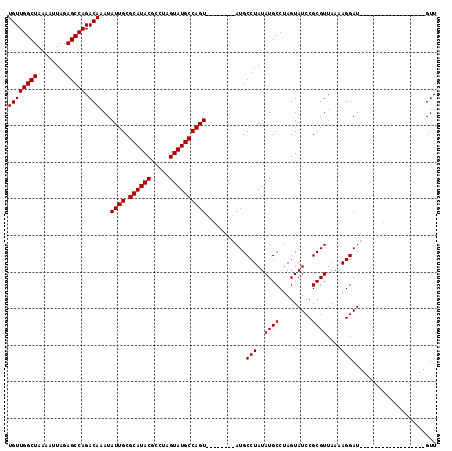
| Location | 23,325,904 – 23,326,012 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23325904 108 + 27905053 UAGGCAUAUAGGCAUACUGGCAUACUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA---UUUUAGCCAACAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCUCGAAC- .((((.....((((...((((((((.....)))))).))....((((.(((((....---....)))))))))..)))).(((.((.........)).))).)))).....- ( -28.80) >DroPse_CAF1 125351 85 + 1 ------------------------CUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA---UUUUAGCCAGCAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCCCGAUCC ------------------------...(((...((.((((...(((.((((((....---....))))))))).))))))(((.((.........)).))).)))....... ( -21.60) >DroSec_CAF1 123210 108 + 1 UAGGCAUAUAGGCAUACUGGCAUACUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA---UUUUAGCCAACAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCCCGAAC- ..(((.....((((...((((((((.....)))))).))....((((.(((((....---....)))))))))..)))).(((.((.........)).))).)))......- ( -27.90) >DroWil_CAF1 131752 87 + 1 ----------------------UACUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAAAUUUUUUAGCCAACAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCUCGA--- ----------------------..(.((((...((.((((...((((.(((((...........))))))))).))))))(((.((.........)).))).)))).).--- ( -19.80) >DroAna_CAF1 119470 83 + 1 ------------------------CUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA---UUUUAGCCAACAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCCC-AAU- ------------------------...(((...((.((((...((((.(((((....---....))))))))).))))))(((.((.........)).))).)))..-...- ( -19.20) >DroPer_CAF1 125811 85 + 1 ------------------------CUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA---UUUUAGCCAGCAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCCCGAUCC ------------------------...(((...((.((((...(((.((((((....---....))))))))).))))))(((.((.........)).))).)))....... ( -21.60) >consensus ________________________CUAGGCGUAUGCGCAAUAUUUGUCUGGCUCUAA___UUUUAGCCAACAAUUUGCCAGCACUUACAUGAUUAAAUUGCAGCCCCGAAC_ ...........................(((...((.((((...((((.(((((...........))))))))).))))))(((.((.........)).))).)))....... (-18.56 = -18.33 + -0.22)



| Location | 23,325,904 – 23,326,012 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23325904 108 - 27905053 -GUUCGAGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGUUGGCUAAAA---UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGUAUGCCUAUAUGCCUA -.....((((.(((.((((......))))))).(((...(((((((((....---....))))).))))((((((.((((((.....))))))))))))))).....)))). ( -35.00) >DroPse_CAF1 125351 85 - 1 GGAUCGGGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGCUGGCUAAAA---UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAG------------------------ ......((((((((.........)))).((((..((((.(((((((((....---....)))))).)))...))))))))..))))..------------------------ ( -23.70) >DroSec_CAF1 123210 108 - 1 -GUUCGGGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGUUGGCUAAAA---UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUAUGCCAGUAUGCCUAUAUGCCUA -.....((((.(((.((((......))))))).(((...(((((((((....---....))))).))))((((((.((((((.....))))))))))))))).....)))). ( -35.00) >DroWil_CAF1 131752 87 - 1 ---UCGAGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGUUGGCUAAAAAAUUUAGAGCCAGACAAAUAUUGCGCAUACGCCUAGUA---------------------- ---...((((((((.........)))).((((..((((.(((((((((...........))))).))))...))))))))..))))....---------------------- ( -20.30) >DroAna_CAF1 119470 83 - 1 -AUU-GGGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGUUGGCUAAAA---UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAG------------------------ -...-.((((((((.........)))).((((..((((.(((((((((....---....))))).))))...))))))))..))))..------------------------ ( -21.30) >DroPer_CAF1 125811 85 - 1 GGAUCGGGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGCUGGCUAAAA---UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAG------------------------ ......((((((((.........)))).((((..((((.(((((((((....---....)))))).)))...))))))))..))))..------------------------ ( -23.70) >consensus _GUUCGGGGCUGCAAUUUAAUCAUGUAAGUGCUGGCAAAUUGUUGGCUAAAA___UUAGAGCCAGACAAAUAUUGCGCAUACGCCUAG________________________ ......((((((((.........)))).((((..((((.(((((((((...........)))))).)))...))))))))..)))).......................... (-21.54 = -21.10 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:35 2006