
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,324,594 – 23,324,726 |
| Length | 132 |
| Max. P | 0.733435 |

| Location | 23,324,594 – 23,324,697 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -30.45 |
| Energy contribution | -30.37 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
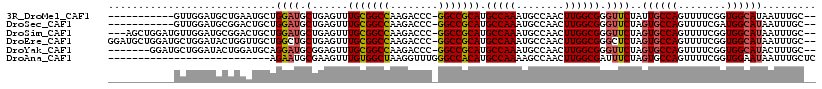
>3R_DroMel_CAF1 23324594 103 - 27905053 UGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAUUGCCAGUUUUCGGUGGCAUAAUUUGC----GUUUUCUAAUUAAAUGCGAUUAGGGGU-A----- (((((((.......-)))))))((((..((((((..(((((((......))))))).......))))))...((((----((((.......)))))))).....)))-)----- ( -37.30) >DroSec_CAF1 121913 103 - 1 UGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGAUGGCAUAAUUUGC----GUUUUCUAAUUAAAUGCGAUUAGGGCG-A----- (((((((.......-)))))))((((((........)))))).((((((((((((........))))))...((((----((((.......)))))))).)))))).-.----- ( -39.90) >DroSim_CAF1 126266 103 - 1 UGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC----GUUUUCUAAUUAAAUGCGAUUAGGGCG-A----- (((((((.......-)))))))((((((........)))))).((((((((((((.(....).))))))...((((----((((.......)))))))).)))))).-.----- ( -39.30) >DroEre_CAF1 128210 103 - 1 UGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGCUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC----GUUUUCUAAUUAAAUGCGAUUAGGGCG-A----- (((((((.......-)))))))((((((........)))))).((((((((((((.(....).))))))...((((----((((.......)))))))).)))))).-.----- ( -41.80) >DroYak_CAF1 133579 103 - 1 UGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUACUUUGC----GUUUUCUAAUUAAAUGCGAUUAGGGCG-A----- (((((((.......-)))))))((((((........)))))).((((((((((((.(....).))))))...((((----((((.......)))))))).)))))).-.----- ( -39.30) >DroAna_CAF1 118179 114 - 1 UGUGGCUAAGGUUUGGGCCACAUGCCAAAAGCCAACUUGGCGAUUUCUAGUGCCAGUUUUCGGUGGAAUAAUUUGCUCUCGUUCUGUAAUUAAAUGCGGUAAGUGAACGCUUUU (((((((........)))))))....((((((..(((((.((.....((((..(((.....((..((....))..))......)))..))))....)).)))))....)))))) ( -28.30) >consensus UGCGGCCAAGACCC_GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC____GUUUUCUAAUUAAAUGCGAUUAGGGCG_A_____ (((((((........)))))))((((((........)))))).((((((((((((........))))))...((((....(((....))).....)))).))))))........ (-30.45 = -30.37 + -0.08)


| Location | 23,324,622 – 23,324,726 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -27.61 |
| Energy contribution | -27.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23324622 104 + 27905053 --GCAAAUUAUGCCACCGAAAACUGGCAAUAGAACCCGCCAAGUUGGCAUUUGGCAUGCGGCC-GGGUCUUGGCCGCAAACUCAGCAUCCAGCAUUCAGCAUCCAAC----------- --((......(((((........)))))...(((...(((((((....))))))).(((((((-(.....))))))))......((.....)).))).)).......----------- ( -32.70) >DroSec_CAF1 121941 104 + 1 --GCAAAUUAUGCCAUCGAAAACUGGCACUAGAACCCGCCAAGUUGGCAUUUGGCAUGCGGCC-GGGUCUUGGCCGCAAACUCAGCAUCCAGCAGUCCGCAUCCAAC----------- --((.....((((((..(..(((((((..........))).))))..)...))))))((((((-(.....))))))).......)).....((.....)).......----------- ( -32.50) >DroSim_CAF1 126294 112 + 1 --GCAAAUUAUGCCACCGAAAACUGGCACUAGAACCCGCCAAGUUGGCAUUUGGCAUGCGGCC-GGGUCUUGGCCGCAAACUCAGCAUCCAGCAGUCCGCAUCCAACAUCCAGCU--- --((......(((((........)))))..............(((((.((.((((.(((((((-(.....))))))))......((.....)).).))).))))))).....)).--- ( -33.20) >DroEre_CAF1 128238 115 + 1 --GCAAAUUAUGCCACCGAAAACUGGCACUAGAGCCCGCCAAGUUGGCAUUUGGCAUGCGGCC-GGGUCUUGGCCGCAAACUCAGCAGCCAGCAACCAGUAUCCAGCAUCCAGCAUCC --(((.....))).........(((((.((.(((...(((((((....))))))).(((((((-(.....))))))))..)))))..))))).............((.....)).... ( -35.60) >DroYak_CAF1 133607 108 + 1 --GCAAAGUAUGCCACCGAAAACUGGCACUAGAACCCGCCAAGUUGGCAUUUGGCAUGCGGCC-GGGUCUUGGCCGCAAACUCCGCAUCCUGCAUCCAGUAUCCAGCAUCC------- --((.......((((........))))(((.((....(((((((....))))))).(((((((-(.....))))))))......((.....)).)).))).....))....------- ( -33.30) >DroAna_CAF1 118215 91 + 1 GAGCAAAUUAUUCCACCGAAAACUGGCACUAGAAAUCGCCAAGUUGGCUUUUGGCAUGUGGCCCAAACCUUAGCCACAAACUUCGCAUUCU--------------------------- ..((...........(((.....))).....(((...((((((......)))))).((((((..........))))))...))))).....--------------------------- ( -20.40) >consensus __GCAAAUUAUGCCACCGAAAACUGGCACUAGAACCCGCCAAGUUGGCAUUUGGCAUGCGGCC_GGGUCUUGGCCGCAAACUCAGCAUCCAGCAGUCAGCAUCCAAC___________ ..((.....((((((..(..(((((((..........))).))))..)...))))))((((((........)))))).......))................................ (-27.61 = -27.67 + 0.06)


| Location | 23,324,622 – 23,324,726 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
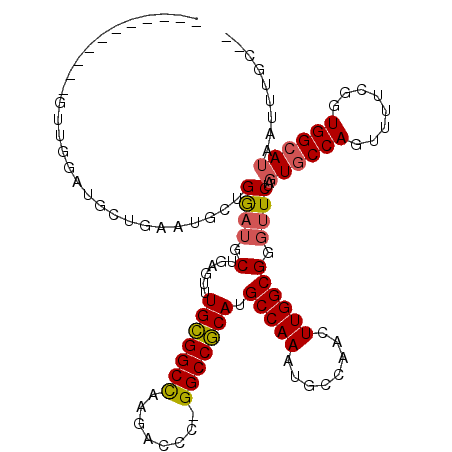
>3R_DroMel_CAF1 23324622 104 - 27905053 -----------GUUGGAUGCUGAAUGCUGGAUGCUGAGUUUGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAUUGCCAGUUUUCGGUGGCAUAAUUUGC-- -----------.......((((((.(((((.....(((..(((((((.......-))))))..(((((........))))))..))).....))))).)))))).(((.....)))-- ( -37.70) >DroSec_CAF1 121941 104 - 1 -----------GUUGGAUGCGGACUGCUGGAUGCUGAGUUUGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGAUGGCAUAAUUUGC-- -----------.((((.((((((((((.....))..)))))))).))))(((((-(((.....)))....(((.....))))))))...((((((........)))))).......-- ( -38.70) >DroSim_CAF1 126294 112 - 1 ---AGCUGGAUGUUGGAUGCGGACUGCUGGAUGCUGAGUUUGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC-- ---.((((((..((((.((((((((((.....))..)))))))).)))).((((-(((.....)))....(((.....)))))))))))))((((.(....).)))).........-- ( -40.90) >DroEre_CAF1 128238 115 - 1 GGAUGCUGGAUGCUGGAUACUGGUUGCUGGCUGCUGAGUUUGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGCUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC-- ....((((((.(((((.((((((.....(.((((..(((((((((((.......-))))))).((.....)).))))..)))).).))))))))))).)))))).(((.....)))-- ( -49.20) >DroYak_CAF1 133607 108 - 1 -------GGAUGCUGGAUACUGGAUGCAGGAUGCGGAGUUUGCGGCCAAGACCC-GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUACUUUGC-- -------....((((((.(((((.((((((((.(((....(((((((.......-)))))))..))....(((.....)))).))))).)))))))).)))))).(((.....)))-- ( -43.80) >DroAna_CAF1 118215 91 - 1 ---------------------------AGAAUGCGAAGUUUGUGGCUAAGGUUUGGGCCACAUGCCAAAAGCCAACUUGGCGAUUUCUAGUGCCAGUUUUCGGUGGAAUAAUUUGCUC ---------------------------.....(((((...(((((((........))))))).(((.(((((.....(((((........)))))))))).))).......))))).. ( -25.10) >consensus ___________GUUGGAUGCUGAAUGCUGGAUGCUGAGUUUGCGGCCAAGACCC_GGCCGCAUGCCAAAUGCCAACUUGGCGGGUUCUAGUGCCAGUUUUCGGUGGCAUAAUUUGC__ ............................((((.(......(((((((........))))))).(((((........)))))).))))..((((((........))))))......... (-28.00 = -28.12 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:32 2006