
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,318,830 – 23,318,972 |
| Length | 142 |
| Max. P | 0.999534 |

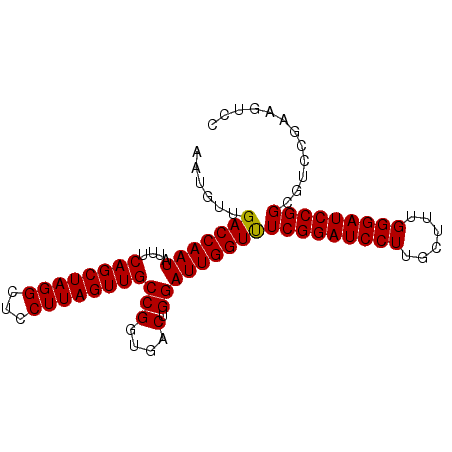
| Location | 23,318,830 – 23,318,920 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -29.34 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318830 90 + 27905053 AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGAUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAUGUCC .(((((((((((...((((((..(((.........)))...).)))))))))).(((((((((......)))))))))....)))))).. ( -28.70) >DroSec_CAF1 116303 90 + 1 AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCC .((((.(((((((....((((((((...))))))))(((....).))))))))).((((((((......))))))))))))......... ( -30.20) >DroSim_CAF1 120559 90 + 1 AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUCUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCC .((((.(((((((....((((((((...))))))))(((....).))))))))).((((((((......))))))))))))......... ( -32.70) >DroEre_CAF1 122550 90 + 1 AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCCUUGGGAUCCGGAGUCGGGAGUCG ...(((((........)))))..(((((((......(((....).))...(..((((((((((......))))))))))..)))))))). ( -36.50) >DroYak_CAF1 127519 90 + 1 AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCCUUGGGAUCCGGAUUCCGUAUUCC ......(((((((....((((((((...))))))))(((....).)))))))))(((((((((......)))))))))............ ( -30.20) >consensus AAUGUUGACCAAUAUUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCC ......(((((((....((((((((...))))))))((((...)))))))))))(((((((((......)))))))))............ (-29.34 = -29.38 + 0.04)

| Location | 23,318,830 – 23,318,920 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318830 90 - 27905053 GGACAUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCAUCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU .(((...((....(((((((........)))))))...))....((((....(((.........)))..))))........)))...... ( -22.50) >DroSec_CAF1 116303 90 - 1 GGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU ((.(....)...))((((((........))))))...((((((.((((....(((.........)))..))))....))))))....... ( -22.80) >DroSim_CAF1 120559 90 - 1 GGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAGACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU ((.(....)...))((((((........))))))..(((((((.((((....(((.........)))..))))....)))))))...... ( -27.20) >DroEre_CAF1 122550 90 - 1 CGACUCCCGACUCCGGAUCCCAAGGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU .............(((((((........)))))))..((((((.((((....(((.........)))..))))....))))))....... ( -22.00) >DroYak_CAF1 127519 90 - 1 GGAAUACGGAAUCCGGAUCCCAAGGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU (((........)))((((((........))))))...((((((.((((....(((.........)))..))))....))))))....... ( -22.30) >consensus GGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAAUAUUGGUCAACAUU .............(((((((........)))))))..((((((.((((....(((.........)))..))))....))))))....... (-22.02 = -22.02 + -0.00)


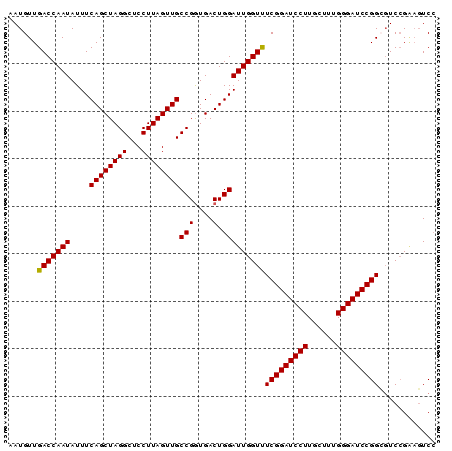
| Location | 23,318,844 – 23,318,947 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -46.72 |
| Consensus MFE | -34.86 |
| Energy contribution | -35.54 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318844 103 + 27905053 UUUCAGCUAGGCUCCUUAGUUGCCGAUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAUGUCC-------------GGAGUCCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))........((((((((..(((((((((......)))))))))...)))))((((-------------(((..........)))))))..))).. ( -37.30) >DroSec_CAF1 116317 110 + 1 UUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC------CGGAGUCCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))..(((..(((((((((..(((((((((......)))))))))...)))..))))))((((------((((..........)))))))))))... ( -45.40) >DroSim_CAF1 120573 110 + 1 UUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUCUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC------CGGAGUCCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))..(((..(((((((((.((((((((((......))))))))).).)))..))))))((((------((((..........)))))))))))... ( -46.70) >DroEre_CAF1 122564 116 + 1 UUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCCUUGGGAUCCGGAGUCGGGAGUCGGGAGUCGGGAGUCGGAGUCCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))..((..(((((((((..((((((((((......))))))))))..))((((..((((.((.(....).)).)))))))))))))).)..))... ( -52.00) >DroYak_CAF1 127533 110 + 1 UUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCCUUGGGAUCCGGAUUCCGUAUUCCGGAGUC------CAGAGACCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))((((....((((..(((((((((((((......)))))))((((((((.....)))))))------).))))))..)))).))))......... ( -52.20) >consensus UUUCAGCUAGGCUCCUUAGUUGCCGGUGACUGGAUUGGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC______CGGAGUCCCUUCCAUCCGGAUUGCCAUA ...((((((((...))))))))..((((((((((((((..(((((((((......)))))))))...))))..................((((....)))).))))).)))))... (-34.86 = -35.54 + 0.68)



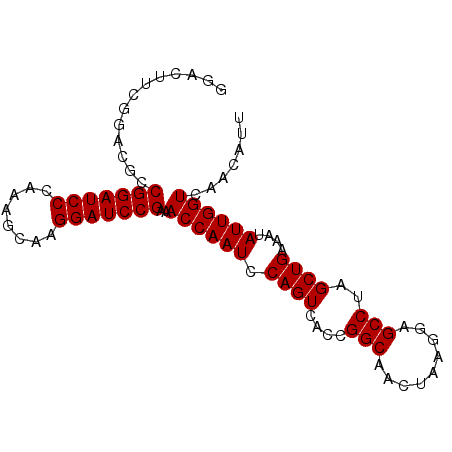
| Location | 23,318,844 – 23,318,947 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -29.18 |
| Energy contribution | -30.18 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318844 103 - 27905053 UAUGGCAAUCCGGAUGGAAGGGACUCC-------------GGACAUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCAUCGGCAACUAAGGAGCCUAGCUGAAA ...(((..(((.(((((...(((.(((-------------(.....))))...(((((((........))))))).......)))..)))))(....)...))))))......... ( -33.30) >DroSec_CAF1 116317 110 - 1 UAUGGCAAUCCGGAUGGAAGGGACUCCG------GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAA ...(((..(((((.((((.((((.((((------(..((((.....))))..))))))))).......((........))..))))....))(....)...))))))......... ( -37.20) >DroSim_CAF1 120573 110 - 1 UAUGGCAAUCCGGAUGGAAGGGACUCCG------GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAGACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAA ...(((..(((((.((((..((.(((.(------(..((((.....))))..))((((((........))))))))).))..))))....))(....)...))))))......... ( -38.60) >DroEre_CAF1 122564 116 - 1 UAUGGCAAUCCGGAUGGAAGGGACUCCGACUCCCGACUCCCGACUCCCGACUCCGGAUCCCAAGGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAA ...(((..(((((((((..((((.((.(....).)).))))............(((((((........)))))))...)).)))).......(....)...))))))......... ( -34.60) >DroYak_CAF1 127533 110 - 1 UAUGGCAAUCCGGAUGGAAGGGUCUCUG------GACUCCGGAAUACGGAAUCCGGAUCCCAAGGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAA ...(((..(((((.((((..(((.((.(------((.((((.....)))).)))((((((........)))))))).)))..))))....))(....)...))))))......... ( -41.20) >consensus UAUGGCAAUCCGGAUGGAAGGGACUCCG______GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACCAAUCCAGUCACCGGCAACUAAGGAGCCUAGCUGAAA ...(((..((((..((((..((...............((((.....))))...(((((((........)))))))...))..))))..)...(....)...))))))......... (-29.18 = -30.18 + 1.00)



| Location | 23,318,880 – 23,318,972 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -23.06 |
| Energy contribution | -24.46 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318880 92 + 27905053 GGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAUGUCC-------------GGAGUCCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGAAGGCGAGUGGAGGAG ((....(((((((......)))))))(((.((((.....)-------------)))))))).(((.((((..(((((..............))))).))))))). ( -33.54) >DroSec_CAF1 116353 99 + 1 GGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC------CGGAGUCCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGAAGGCGUGUGGAGGAG ....(((((((((......)))))))))(.((((.((((((((...------.((((....)))).))))))))(((..............)))...)))).).. ( -36.84) >DroSim_CAF1 120609 99 + 1 GGUCUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC------CGGAGUCCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGAAGGCGAGUGGAGGAG ((.((((((((((......)))))))((..((((.....))))..)------).))).))..(((.((((..(((((..............))))).))))))). ( -39.24) >DroEre_CAF1 122600 104 + 1 GGUUUCGGAUCCUUGCCUUGGGAUCCGGAGUCGGGAGUCGGGAGUCGGGAGUCGGAGUCCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGGAGGCAAGU-GGGGUG (..((((((((((......))))))))))..).((((..((((.((.(....).)).))))))))((((.(.(((((..............))))).)-.)))). ( -42.84) >DroYak_CAF1 127569 94 + 1 GGUUUCGGAUCCUUGCCUUGGGAUCCGGAUUCCGUAUUCCGGAGUC------CAGAGACCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGGAGU----G-GGAGUG (((((((((((((......)))))))((((((((.....)))))))------).))))))(((((((((.((((......))))....))).)----)-)))).. ( -44.80) >consensus GGUUUCGGAUCCUUGCUUUGGGAUCCGGCGUCCGAAGUCCGGAGUC______CGGAGUCCCUUCCAUCCGGAUUGCCAUAAAUUAUGAGAAGGCGAGUGGAGGAG (((.(((((((((......))))))))).......((((((((..........((((....)))).)))))))))))............................ (-23.06 = -24.46 + 1.40)

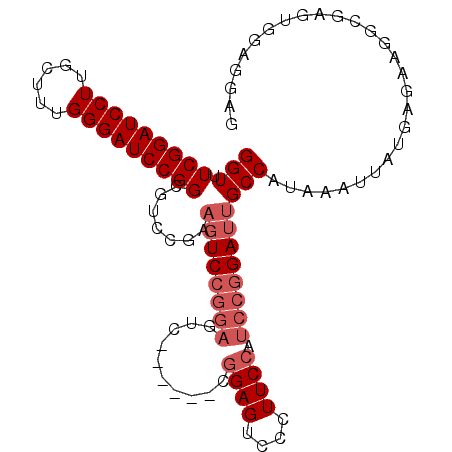

| Location | 23,318,880 – 23,318,972 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -19.34 |
| Energy contribution | -20.58 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23318880 92 - 27905053 CUCCUCCACUCGCCUUCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGACUCC-------------GGACAUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACC ....(((....(((..............)))..((((((.(.......))))-------------)))....)))...(((((((........)))))))..... ( -27.64) >DroSec_CAF1 116353 99 - 1 CUCCUCCACACGCCUUCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGACUCCG------GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACC .(((((((..((..((.(((((...))))).))..))..)))).((((.((((------(..((((.....))))..))))))))).......)))......... ( -32.70) >DroSim_CAF1 120609 99 - 1 CUCCUCCACUCGCCUUCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGACUCCG------GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAGACC .(((((((..((..((.(((((...))))).))..))..)))).((((.((((------(..((((.....))))..))))))))).......)))......... ( -33.50) >DroEre_CAF1 122600 104 - 1 CACCCC-ACUUGCCUCCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGACUCCGACUCCCGACUCCCGACUCCCGACUCCGGAUCCCAAGGCAAGGAUCCGAAACC ......-.(((((((............(((..(((((((.(((.((((.((.(....).)).))))...)))....))))))).))))))))))........... ( -31.50) >DroYak_CAF1 127569 94 - 1 CACUCC-C----ACUCCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGUCUCUG------GACUCCGGAAUACGGAAUCCGGAUCCCAAGGCAAGGAUCCGAAACC ..((.(-(----(.((((((((...)))))......)))))).))(((.((.(------((.((((.....)))).)))((((((........)))))))).))) ( -29.70) >consensus CUCCUCCACUCGCCUUCUCAUAAUUUAUGGCAAUCCGGAUGGAAGGGACUCCG______GACUCCGGACUUCGGACGCCGGAUCCCAAAGCAAGGAUCCGAAACC ...........(((..............)))..((((((((((......)))).........))))))..........(((((((........)))))))..... (-19.34 = -20.58 + 1.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:26 2006