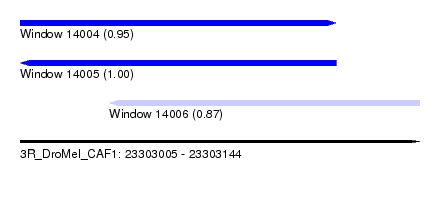
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,303,005 – 23,303,144 |
| Length | 139 |
| Max. P | 0.997184 |

| Location | 23,303,005 – 23,303,115 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23303005 110 + 27905053 CUGUUUUUUUUUUAUAUAAA-----ACAGCAAGUU----CUGCAGGGUAUCAAAACUGCUUUGUCGCAAAGCAUACUUUUGCAUGUGCCAAAGCAAUUGUACAGUGCUCGCCCAGCAAA (((((((..........)))-----))))...(((----..((.((((((...((.(((((((.((((..(((......))).)))).))))))).)).....))))))))..)))... ( -27.90) >DroSec_CAF1 100414 105 + 1 ----UUUUGUUUGAUAUGGA-----ACAGCAAGUU----CUGCAGGGUAUCAAAACUGCUUU-UAGCAAAGCAUGCUUUUGCAUGUGCCAAAGCAAUUGUACAGUGCUUUCCCAGCAAA ----.((((((.(....(((-----((.....)))----))(((..(((.(((...((((((-..((...((((((....)))))))).)))))).))))))..)))....).)))))) ( -26.90) >DroSim_CAF1 104284 83 + 1 ----UUU---------------------------U----CUGCAGGGUAUCAAAACUGCUUU-UCGCAAAGCAUGCUUUUGCAUGUGCCAAAGCAAUUGUACAGUGCUUUCCCAGCAAA ----...---------------------------.----..(((..(((.(((...((((((-..((...((((((....)))))))).)))))).))))))..)))............ ( -21.20) >DroEre_CAF1 106862 108 + 1 ----CU----UCGAUAUAAAAAU--ACAGCAAGUUGGUGCUGCAGGGUAUCAAAACUGCUUU-UCGCAAAGCAUGCUGUGGCCUGUGCCAAAGCAAUUGUACAGUGCUUUCCCAGCAAA ----..----..(((((......--.(((((......)))))....))))).....((((((-....))))))(((((.(((((((((..((....)))))))).)))....))))).. ( -29.70) >DroYak_CAF1 110803 114 + 1 ----CUUUGUUUAAUAGAAAAAUAAACAGUAAGUUAUUUCAGCAGGGUAUCAAAGCUGCUUU-UCGCAAAGCAUGCUUUGGCAUGUGCCAAAGCAAUUGUACAGUGCUUUCCCAGCAAA ----..(((((((.........)))))))............((.(((....(((((((((((-....))))))(((((((((....)))))))))..........)))))))).))... ( -31.90) >consensus ____UUUU_UUUGAUAUAAA_____ACAGCAAGUU____CUGCAGGGUAUCAAAACUGCUUU_UCGCAAAGCAUGCUUUUGCAUGUGCCAAAGCAAUUGUACAGUGCUUUCCCAGCAAA .........................................(((..(((.(((...((((((...((...((((((....)))))))).)))))).))))))..)))............ (-18.88 = -19.28 + 0.40)


| Location | 23,303,005 – 23,303,115 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23303005 110 - 27905053 UUUGCUGGGCGAGCACUGUACAAUUGCUUUGGCACAUGCAAAAGUAUGCUUUGCGACAAAGCAGUUUUGAUACCCUGCAG----AACUUGCUGU-----UUUAUAUAAAAAAAAAACAG (((((.(((.......(((..((((((((((((((((((....)))))...)))..))))))))))...)))))).))))----).....((((-----(((..........))))))) ( -27.91) >DroSec_CAF1 100414 105 - 1 UUUGCUGGGAAAGCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCUA-AAAGCAGUUUUGAUACCCUGCAG----AACUUGCUGU-----UCCAUAUCAAACAAAA---- ((((.(((((.((((.........(((((((((((((((....)))))...)))).-))))))((((((........)))----))).)))).)-----))))...)))).....---- ( -30.00) >DroSim_CAF1 104284 83 - 1 UUUGCUGGGAAAGCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCGA-AAAGCAGUUUUGAUACCCUGCAG----A---------------------------AAA---- (((((.((((((((..........((((((.((....)).))))))((((((....-)))))))))))....))).))))----)---------------------------...---- ( -26.00) >DroEre_CAF1 106862 108 - 1 UUUGCUGGGAAAGCACUGUACAAUUGCUUUGGCACAGGCCACAGCAUGCUUUGCGA-AAAGCAGUUUUGAUACCCUGCAGCACCAACUUGCUGU--AUUUUUAUAUCGA----AG---- ..(((((..((((((.(.....).))))))(((....))).)))))((((((....-))))))..(((((((...(((((((......))))))--)......))))))----).---- ( -32.90) >DroYak_CAF1 110803 114 - 1 UUUGCUGGGAAAGCACUGUACAAUUGCUUUGGCACAUGCCAAAGCAUGCUUUGCGA-AAAGCAGCUUUGAUACCCUGCUGAAAUAACUUACUGUUUAUUUUUCUAUUAAACAAAG---- ...((.((((((((..........(((((((((....)))))))))((((((....-)))))))))))....))).)).............((((((.........))))))...---- ( -34.30) >consensus UUUGCUGGGAAAGCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCGA_AAAGCAGUUUUGAUACCCUGCAG____AACUUGCUGU_____UUUAUAUCAAA_AAAA____ .((((.((((((((..........((((((.((....)).))))))((((((.....)))))))))))....))).))))....................................... (-22.08 = -22.20 + 0.12)



| Location | 23,303,036 – 23,303,144 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -18.85 |
| Energy contribution | -21.53 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23303036 108 - 27905053 CUUCCAGUUUGGCGGUAAAUCAUCAC-----------CGUUUUGCUGGGCGA-GCACUGUACAAUUGCUUUGGCACAUGCAAAAGUAUGCUUUGCGACAAAGCAGUUUUGAUACCCUGCA ..((((((..((((((........))-----------))))..))))))...-(((..((((((((((((((((((((((....)))))...)))..)))))))))..)).)))..))). ( -37.10) >DroSec_CAF1 100441 107 - 1 CUUCCAGUUUGGCGGUAAAUCAUCAC-----------CGUUUUGCUGGGAAA-GCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCUA-AAAGCAGUUUUGAUACCCUGCA .(((((((..((((((........))-----------))))..)))))))..-(((..((((((((((((((((((((((....)))))...)))).-))))))))..)).)))..))). ( -40.60) >DroSim_CAF1 104289 107 - 1 CUUCCAGUUUGGCGGUAAAUCAUCAC-----------CGUUUUGCUGGGAAA-GCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCGA-AAAGCAGUUUUGAUACCCUGCA .(((((((..((((((........))-----------))))..)))))))..-(((..(((((((((((((.((((((((....)))))...)))..-))))))))..)).)))..))). ( -39.70) >DroEre_CAF1 106892 107 - 1 CUUCCAGUUUGGCGGUAAAUCAUCGC-----------CGUUUUGCUGGGAAA-GCACUGUACAAUUGCUUUGGCACAGGCCACAGCAUGCUUUGCGA-AAAGCAGUUUUGAUACCCUGCA .(((((((.(((((((.....)))))-----------))....)))))))..-(((..((((((.((((.((((....)))).))))((((((....-))))))...))).)))..))). ( -40.80) >DroYak_CAF1 110839 107 - 1 CUUCCAGUUUGGCGGUAAAUCAUCAC-----------CGUUUUGCUGGGAAA-GCACUGUACAAUUGCUUUGGCACAUGCCAAAGCAUGCUUUGCGA-AAAGCAGCUUUGAUACCCUGCU .(((((((..((((((........))-----------))))..))))))).(-(((..((((((.(((((((((....)))))))))((((((....-))))))...))).)))..)))) ( -44.40) >DroAna_CAF1 97417 102 - 1 CUUGCAUUUUGGCGGUAAAUCAUCGCACACAGCCACACAAUUUGCUCAGAAAUGCACACUACAAUUGCGU----AAAU------GCAUCACCU-------ACCAGUUUUG-UAUCCUGGA ..((((((((((((((.....))))).)).....((.((((((((........)))......))))).))----))))------)))......-------.((((.....-....)))). ( -19.50) >consensus CUUCCAGUUUGGCGGUAAAUCAUCAC___________CGUUUUGCUGGGAAA_GCACUGUACAAUUGCUUUGGCACAUGCAAAAGCAUGCUUUGCGA_AAAGCAGUUUUGAUACCCUGCA .(((((((..((((.......................))))..)))))))...(((..(((((((((((((.((((((((....)))))...)))...))))))))..)).)))..))). (-18.85 = -21.53 + 2.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:05 2006