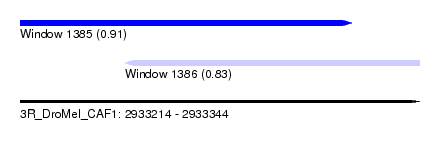
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,933,214 – 2,933,344 |
| Length | 130 |
| Max. P | 0.913600 |

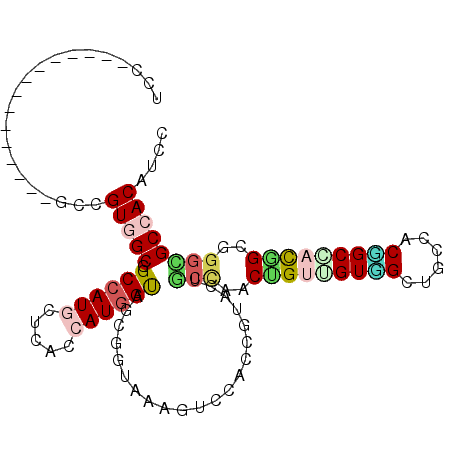
| Location | 2,933,214 – 2,933,322 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.33 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.55 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2933214 108 + 27905053 UCGGCGGACUCAGCCUCGGCGGUGGCCGCCAUGCUCACCAUGGCAGCGGUAAAGUCCACCAGAGCUGCGCCCGAAGUGGCUGCCACUGCUACGGCCGGAGCCACGUCC ..(((((.(((.(((..(((((((((.(((((..((.....(((.(((((.............)))))))).)).))))).)))))))))..)))..))).).)))). ( -48.62) >DroVir_CAF1 14946 93 + 1 UCC---------------GCCGUAGCGGCCAUGCUCACCAUGGUGGCCGUAAAGUCCAGCGUGCUGCUAACUGUGGUCGCCGCAACGGCCACAGCGGGCGCCACGUCC ...---------------......((((((((..........))))))))........(((((.((((..(((((((((......)))))))))..)))).))))).. ( -40.70) >DroPse_CAF1 12959 93 + 1 UCC---------------GCUGUGGCGGCCAUGGAGACCAUUGUGGCGGUAAAGUCCACGGUAGUGCCUACGGUAGUGGCGGCUACCGCCAUGGCUGGUGCCACAUCC ...---------------..((((((((((((((........(((((......).))))(((((((((.((....))))).)))))).))))))))...))))))... ( -44.20) >DroGri_CAF1 20469 93 + 1 UCC---------------GCCGUGGCGGCCAUAUUCACCAUGGUCGCUGUAAAAUCCAAAGUGCUGCCAACUGUUGUGGCUGCCACGGCAACAGCGGGCGCCACAUCC ...---------------(((((((((((((((..((...((((((((...........))))..))))..)).)))))))))))))))....((....))....... ( -39.40) >DroEre_CAF1 13449 105 + 1 UCGGCGGACUCAGCCUCAGCGGUGGCCGCCAUGCUCACCAUGGCAGCGGUAAAGUCCACCAGAGAGC---CCGAUGUGGCUGCCACUGCAACGGCCGGAGCCACGUCC ..(((((.(((.(((...((((((((.(((((((((....(((..((......))...)))..))))---.....))))).))))))))...)))..))).).)))). ( -44.40) >DroMoj_CAF1 20006 93 + 1 UCC---------------GCCGUAGCGGCCAUAUUCACCAUGGUCGCCGUAAAGUCCAGCGUGCUGCUAACUGUUGUUGCUGCCACGGCCACGGCGGGCGCCACAUCC (((---------------(((((...((((((.......))))))(((((......(((((.((.((.....)).)))))))..))))).)))))))).......... ( -34.70) >consensus UCC_______________GCCGUGGCGGCCAUGCUCACCAUGGUAGCGGUAAAGUCCACCGUACUGCCAACUGUUGUGGCUGCCACGGCCACGGCGGGCGCCACAUCC .....................(((((.((((((.....)))))).....................(((..(((((((((......)))))))))..)))))))).... (-25.08 = -25.55 + 0.47)

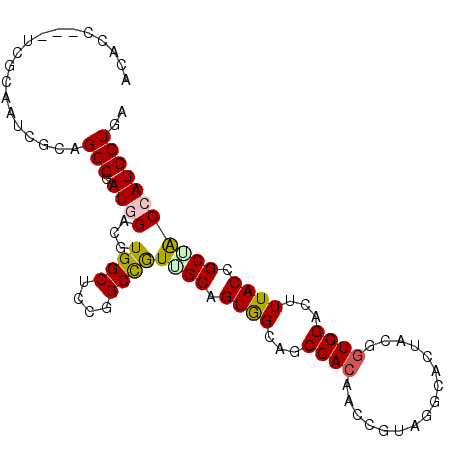
| Location | 2,933,248 – 2,933,344 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -20.20 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2933248 96 - 27905053 ACACC---UCGCAAUCGCAGCCGAUGGACGUGGCUCCGGCCGUAGCAGUGGCAGCCACUUCGGGCGCAGCUCUGGUGGACUUUACCGCUGCCAUGGUGA .....---.......((.(((((.......))))).))(((((.(((((((.(((((((..((((...)))).))))).))...))))))).))))).. ( -37.00) >DroPse_CAF1 12978 99 - 1 ACACCACAUCCCAAUCACAGCCCAUGGAUGUGGCACCAGCCAUGGCGGUAGCCGCCACUACCGUAGGCACUACCGUGGACUUUACCGCCACAAUGGUCU ...((((((((..............)))))))).((((....(((((((((...((((....((((...)))).))))...)))))))))...)))).. ( -37.04) >DroGri_CAF1 20488 87 - 1 ACGCC------------CAGCCAAUGGAUGUGGCGCCCGCUGUUGCCGUGGCAGCCACAACAGUUGGCAGCACUUUGGAUUUUACAGCGACCAUGGUGA .....------------..((((((.(.((((((((((((....).)).))).)))))).).))))))..((((.(((............))).)))). ( -32.90) >DroEre_CAF1 13483 93 - 1 ACACC---UCGCAGUCGCAGCCGAUGGACGUGGCUCCGGCCGUUGCAGUGGCAGCCACAUCGG---GCUCUCUGGUGGACUUUACCGCUGCCAUGGUGA .((((---..(((((((.(((((.......))))).))))...))).((((((((...(((((---.....)))))((......)))))))))))))). ( -37.50) >DroYak_CAF1 12835 96 - 1 ACACC---UCGCAGUCGCAGCCGAUGGACGUGGCUCCGGCCGUUGCAGUGGCAGCCACAUCUGGCGGCGCUCUGGUGGAUUUUACCGCUGCCAUGGUGA .((((---..(((((.((.((((..(((.((((((...((((......)))))))))).)))..))))))...((((.....)))))))))...)))). ( -43.10) >DroPer_CAF1 13020 99 - 1 ACACCACAUCCCAAUCACAGCCCAUGGAUGUGGCACCCGCCAUGGCGGUAGCCGCCACUACCGUAGGCACUACCGUGGACUUUACCGCCACAAUGGUCU ...((((((((..............)))))))).....(((((((((((((...((((....((((...)))).))))...)))))))))...)))).. ( -36.14) >consensus ACACC___UCGCAAUCGCAGCCGAUGGACGUGGCUCCGGCCGUUGCAGUGGCAGCCACAACCGUAGGCACUACGGUGGACUUUACCGCUACCAUGGUGA ...................(((.((((...((((....))))((((.((((...((((................))))...)))).))))))))))).. (-20.20 = -19.92 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:15 2006