
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
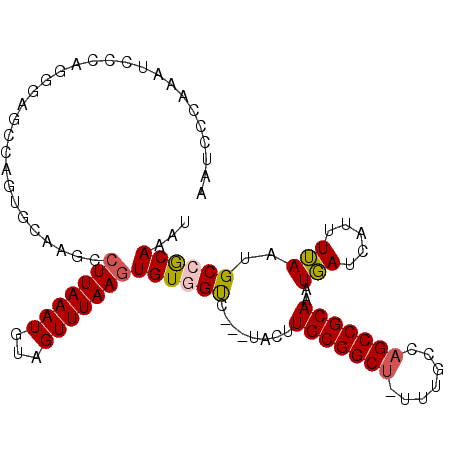
| Location | 23,297,303 – 23,297,408 |
| Length | 105 |
| Max. P | 0.938189 |

| Location | 23,297,303 – 23,297,408 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23297303 105 + 27905053 AAUCGCAAAUCCCAGGGAGCUAGUGCCAGCCUUAAAUGUAGUUUAAUUGUGGUAUACUACUUGCGGCU-UUUACCAGCCGCAAAUGAUCAUUUUAAUGCCGCAAAU .............((((.((....))...)))).............(((((((((.....((((((((-......)))))))).(((.....)))))))))))).. ( -27.10) >DroPse_CAF1 99049 102 + 1 AAUCCUAAAUGCGGCAUGUCCAGGGCAAUCCUUAAAUGCAGUUUAAGUGUUGUU---UACCUGCGGCU-UUUGGCAGCCGCAAAUGAUCCUUUUAAUGCCGCAAAU .........((((((((....(((((((..(((((((...)))))))..)))).---....(((((((-......)))))))......)))....))))))))... ( -37.10) >DroSec_CAF1 94885 105 + 1 AAUCGCAAAUCCCAGUGAGAUUGUGCCAGCCUUAAAUGUAGUUUAAGUGUGGUCUACUACUUGCGGCU-UUUGCCAGCCGCAAAUGAUCAUUUUAAUGCCGCAAAU ..((((........))))(((..(((((..(((((((...)))))))..)))).......((((((((-......)))))))))..)))................. ( -27.50) >DroEre_CAF1 100440 105 + 1 AAUCGCAAAUCCCAGGGCGCCAGUGCCAGCCUUAAAUGUAGUUUAAGUGUGGUCUACUACUUGCGGCA-UUUGCCCGCCGCAAAUGAUCAUUUUAGUGCCGCAAAU ...............(((((.(((((((..(((((((...)))))))..))))..)))..(((((((.-.......)))))))............)))))...... ( -29.90) >DroAna_CAF1 90897 97 + 1 AAUCCAAAAUCUGAGGC------UGGAAUCCUUAAAUGUAGUUUAAGUGUGGCC---CAUUUGCGGCUUUUUGCCAGCCGCAAAUUAUCCUUUAAAUGCCACAAAU ..((((...........------))))...(((((((...)))))))((((((.---..(((((((((.......)))))))))(((.....)))..))))))... ( -25.70) >DroPer_CAF1 99484 102 + 1 AAUCCUAAAUGCGGCGUGUCCAGGGCAAUCCUUAAAUGCAGUUUAAGUGUUGUU---UACCUGCGGCU-UUUGGCAGCCGCAAAUGAUCCUUUUAAUGCCGCAAAU .........((((((((....(((((((..(((((((...)))))))..)))).---....(((((((-......)))))))......)))....))))))))... ( -36.70) >consensus AAUCCCAAAUCCCAGGGAGCCAGUGCAAGCCUUAAAUGUAGUUUAAGUGUGGUC___UACUUGCGGCU_UUUGCCAGCCGCAAAUGAUCAUUUUAAUGCCGCAAAU ..............................(((((((...)))))))((((((........(((((((.......)))))))..(((.....)))..))))))... (-17.24 = -17.35 + 0.11)


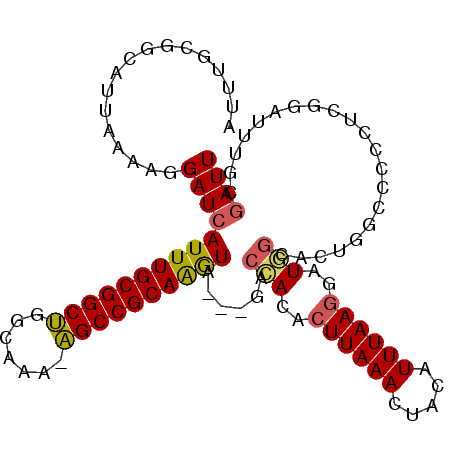
| Location | 23,297,303 – 23,297,408 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23297303 105 - 27905053 AUUUGCGGCAUUAAAAUGAUCAUUUGCGGCUGGUAAA-AGCCGCAAGUAGUAUACCACAAUUAAACUACAUUUAAGGCUGGCACUAGCUCCCUGGGAUUUGCGAUU .....((((.(((((........((((((((......-))))))))(((((.((.......)).))))).))))).))))(((((((....))))....))).... ( -28.40) >DroPse_CAF1 99049 102 - 1 AUUUGCGGCAUUAAAAGGAUCAUUUGCGGCUGCCAAA-AGCCGCAGGUA---AACAACACUUAAACUGCAUUUAAGGAUUGCCCUGGACAUGCCGCAUUUAGGAUU ...((((((((....(((...((((((((((......-)))))))))).---..(((..((((((.....))))))..))).)))....))))))))......... ( -38.00) >DroSec_CAF1 94885 105 - 1 AUUUGCGGCAUUAAAAUGAUCAUUUGCGGCUGGCAAA-AGCCGCAAGUAGUAGACCACACUUAAACUACAUUUAAGGCUGGCACAAUCUCACUGGGAUUUGCGAUU .....((((.(((((......((((((((((......-)))))))))).((((............)))).))))).))))(((.(((((.....)))))))).... ( -32.20) >DroEre_CAF1 100440 105 - 1 AUUUGCGGCACUAAAAUGAUCAUUUGCGGCGGGCAAA-UGCCGCAAGUAGUAGACCACACUUAAACUACAUUUAAGGCUGGCACUGGCGCCCUGGGAUUUGCGAUU (((((((((.(......)...((((((.....)))))-)))))))))).(((((((.((((((((.....))))))...(((......))).)))).))))).... ( -30.00) >DroAna_CAF1 90897 97 - 1 AUUUGUGGCAUUUAAAGGAUAAUUUGCGGCUGGCAAAAAGCCGCAAAUG---GGCCACACUUAAACUACAUUUAAGGAUUCCA------GCCUCAGAUUUUGGAUU (((((.(((.......((...((((((((((.......)))))))))).---..))...((((((.....)))))).......------))).)))))........ ( -31.20) >DroPer_CAF1 99484 102 - 1 AUUUGCGGCAUUAAAAGGAUCAUUUGCGGCUGCCAAA-AGCCGCAGGUA---AACAACACUUAAACUGCAUUUAAGGAUUGCCCUGGACACGCCGCAUUUAGGAUU ...((((((......(((...((((((((((......-)))))))))).---..(((..((((((.....))))))..))).)))......))))))......... ( -35.10) >consensus AUUUGCGGCAUUAAAAGGAUCAUUUGCGGCUGGCAAA_AGCCGCAAGUA___GACCACACUUAAACUACAUUUAAGGAUGGCACUGGCCCCCUCGGAUUUGCGAUU .................((((((((((((((.......))))))))))......(((..((((((.....))))))..))).....................)))) (-19.40 = -19.10 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:56 2006