
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,296,732 – 23,296,872 |
| Length | 140 |
| Max. P | 0.918305 |

| Location | 23,296,732 – 23,296,836 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -33.49 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.39 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
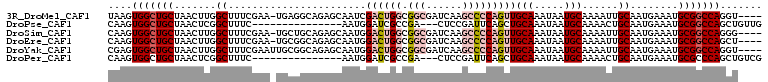
>3R_DroMel_CAF1 23296732 104 - 27905053 UAAGUGGCUGCUAACUUGGCUUUCGAA-UGAGGCAGAGCAAUCGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAAUGCAAAAUUGCAAUGAAAUGCGGCCAGGU---- ....(((((((....(((.(((.....-.))).))).(((((.((((((.(((......)))))))))(((.....)))...)))))........)))))))...---- ( -37.90) >DroPse_CAF1 98439 91 - 1 CAAGUGGCUGCUAACUCGGCUUUC---------------AAUGGAUCGCCGA---CUCCGAUUCAGCUGCAAAUAAUGCAAAACUGCAAUGAAAUGCGGCCAGCUGUUG ..((..((((.....(((((..((---------------....))..)))))---..........((((((..((.((((....)))).))...))))))))))..)). ( -28.20) >DroSim_CAF1 98286 104 - 1 CAAGUGGCUGCUAACUUGGCUUUCGAA-UGCUGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAAUGCAAAAUUGCAAUGAAAUGCGGCCAGGG---- ....(((((((......(((.......-.))).((..(((((.((((((.(((......)))))))))(((.....)))...)))))..))....)))))))...---- ( -37.60) >DroEre_CAF1 99899 104 - 1 CAAGUGGCUGCUAACUUGGCUUUCGAA-UGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAAUGCAAAAUUGCAAUGAAAUGCGGCCAGCU---- ....(((((((.......((.......-.))..((..(((((.((((((.(((......)))))))))(((.....)))...)))))..))....)))))))...---- ( -36.80) >DroYak_CAF1 97213 105 - 1 CGAGUGGCUGCUAACUUGGCUUUCGAAUUGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAAUGCAAAAUUGCAAUGAAAUGCGGCCAGGU---- ....(((((((.......((......(((((......))))).((((((.(((......)))))))))))......((((....)))).......)))))))...---- ( -36.80) >DroPer_CAF1 98873 91 - 1 CAAGUGGCUGCUAACUCGGCUUUC---------------AAUGGAUCGCCGA---CUCCGAUUCAGCUGCAAAUAAUGCAAAACUGCAAUGAAAUGCGCCCAGCUGUCG ...(..((((.....(((((..((---------------....))..)))))---........))))..)......((((....))))..((...((.....))..)). ( -23.62) >consensus CAAGUGGCUGCUAACUUGGCUUUCGAA_UGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAAUGCAAAAUUGCAAUGAAAUGCGGCCAGCU____ ....(((((((.......((.......................((((((.(((......)))))))))(((.....)))......))........)))))))....... (-21.00 = -22.39 + 1.39)
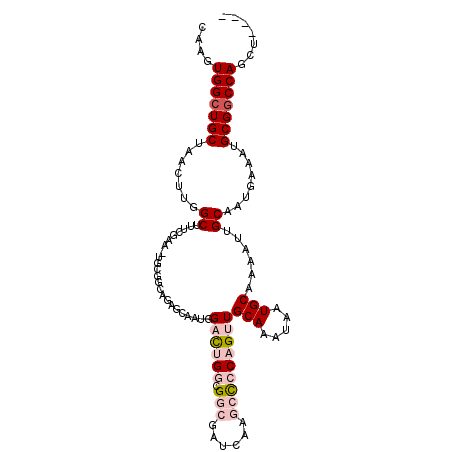
| Location | 23,296,761 – 23,296,872 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23296761 111 - 27905053 ACUCUUG----UUGGCCCGGCUAAUAGCAGAAAGCAAACUUAAGUGGCUGCUAACUUGGCUUUCGAA-UGAGGCAGAGCAAUCGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA .((((((----..((((.((....((((((...((........))..)))))).)).))))..))).-.)))((.((....))((((((.(((......)))))))))))...... ( -37.80) >DroSec_CAF1 94347 111 - 1 ACUCAUG----UUGGCCCGGCUAAUAGCAGCAAGCAAACUCAAGUGGCUGCUAACUUGGCUUUCGAA-UGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA ...((((----((.(((((((((((((((((..((........)).)))))))..))))))......-.).)))..))).)))((((((.(((......)))))))))........ ( -36.11) >DroSim_CAF1 98315 111 - 1 ACUCAUG----UUGGCCCGGCUAAUAGCAGCAAGCAAACUCAAGUGGCUGCUAACUUGGCUUUCGAA-UGCUGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA .(((.((----(((((...)))))))((((((..((((..(((((........)))))..))).)..-)))))).))).....((((((.(((......)))))))))........ ( -39.40) >DroEre_CAF1 99928 111 - 1 GGCACUG----UUGGCCCGGCUAAUAGCGGCAAGCAAACUCAAGUGGCUGCUAACUUGGCUUUCGAA-UGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA .((.(((----(((.(..(((((((((((((..((........)).)))))))..))))))...)..-..)))))).))....((((((.(((......)))))))))........ ( -38.00) >DroYak_CAF1 97242 112 - 1 ACUUCUG----UUGGCUUGGCUAAUAGCAGCAAGCAAACUCGAGUGGCUGCUAACUUGGCUUUCGAAUUGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA ...((((----((((((((((.....))..)))))....(((((.((((........)))))))))....)))))))(((....(((((.(((......)))))))))))...... ( -38.40) >DroPer_CAF1 98906 97 - 1 GCCUCUGCUACUGAGGAGGGCUAAUGGGAGCAAGCA-ACUCAAGUGGCUGCUAACUCGGCUUUC---------------AAUGGAUCGCCGA---CUCCGAUUCAGCUGCAAAUAA .....(((..((((((((.......(..((((.((.-((....)).))))))..)(((((..((---------------....))..)))))---))))...))))..)))..... ( -30.90) >consensus ACUCCUG____UUGGCCCGGCUAAUAGCAGCAAGCAAACUCAAGUGGCUGCUAACUUGGCUUUCGAA_UGCGGCAGAGCAAUGGACUGGCGGCGAUCAAGCCCCAGUUGCAAAUAA ..............((..(((((((((((((..((........)).)))))))..))))))......................((((((.(((......)))))))))))...... (-23.82 = -24.77 + 0.95)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:55 2006