| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,288,670 – 23,288,798 |
| Length | 128 |
| Max. P | 0.969519 |

| Location | 23,288,670 – 23,288,777 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.41 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -13.79 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
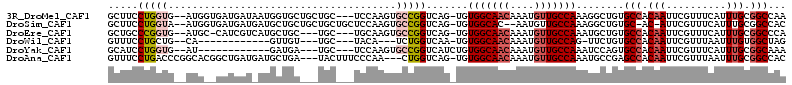
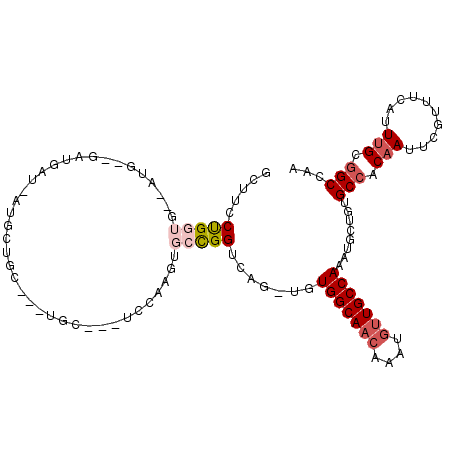
>3R_DroMel_CAF1 23288670 107 + 27905053 GCUUCCUGGUG--AUGGUGAUGAUAAUGGUGCUGCUGC---UCCAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAGGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAA ......(((((--((((.(((((...(((.((....))---.))).(((((((((..-..(((((((....)))))))..))))).).)))...))))))))))....)))). ( -35.10) >DroSim_CAF1 89436 106 + 1 GCUUCCUGGUA--AUGGUGAUGAUGAUGCUGCUGCUGCUGCUCCAAGUGCCGGUCAG-UGUGGCAC--AAUGUUGCCAAAGGCUGUGC-AC-AUUCGUUUCAUUUGCGGCCAC ......((((.--..(((((.(((((.((.((....)).)).....(((((((((..-..(((((.--.....)))))..))))).))-))-..))))))))))....)))). ( -34.80) >DroEre_CAF1 91759 103 + 1 GCUGCCCGGUG--AUGC-CAUCGUCAUGCUGC---UGC---UGCAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCCA ((.((.(((((--....-)))))....)).))---.((---((((((((..((((((-(.(((((((....)))))))...)))).)))((.....))..))))))))))... ( -39.10) >DroWil_CAF1 94398 88 + 1 GUUUCCUGCUG--CA------------GUUGU---UGC---UACA---UCUGGUCAA-UGUGGCAACAAAUGUUGCCAG-UUCUGUGCCACAAUUCGUUUAAUUUGUGGCUAG .....((((.(--((------------.((((---(((---((((---(.......)-))))))))))).))).).)))-..(((.(((((((..........)))))))))) ( -31.00) >DroYak_CAF1 89048 93 + 1 GCAUCCUGGUG--AU------------GAUGA---UGC---UCCAAGUGCCGGUCAUCUGUGGCAACAAAUGUUGCCAAAUCCAGUGCCACAAUUCGUUUCAUUUGCGGCAAA .....((((.(--((------------(((..---.((---.......))..))))))..(((((((....)))))))...))))((((.(((..........))).)))).. ( -29.60) >DroAna_CAF1 82014 106 + 1 GUUUCCUGACCCGGCACGGCUGAUGAUGCUGA---UACUUUCCCAA---CUGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCCGAGCCACAAUUCGUUUAAUUUGCGGCCAC ...........(((((((((.......))))(---((((..((...---..))..))-)))((((((....))))))...))))).(((.(((..........))).)))... ( -32.70) >consensus GCUUCCUGGUG__AUG__GAUGAU_AUGCUGC___UGC___UCCAAGUGCCGGUCAG_UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAA .....(((((......................................))))).......(((((((....)))))))........(((.(((..........))).)))... (-13.79 = -14.76 + 0.97)

| Location | 23,288,670 – 23,288,777 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.41 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23288670 107 - 27905053 UUGGCCGCAAAUGAAACGAAUUGUGGCACAGCCUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUGGA---GCAGCAGCACCAUUAUCAUCACCAU--CACCAGGAAGC .(((((((((..........))))))).))(((..(((((((....)))))))..-......)))...(((.---((....)).))).........((..--.....)).... ( -27.20) >DroSim_CAF1 89436 106 - 1 GUGGCCGCAAAUGAAACGAAU-GU-GCACAGCCUUUGGCAACAUU--GUGCCACA-CUGACCGGCACUUGGAGCAGCAGCAGCAGCAUCAUCAUCACCAU--UACCAGGAAGC (((((((((.((.......))-.)-)).(((....(((((.....--.)))))..-)))...)))...((..((.((....)).))..))....)))...--........... ( -22.50) >DroEre_CAF1 91759 103 - 1 UGGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUGCA---GCA---GCAGCAUGACGAUG-GCAU--CACCGGGCAGC ((((((((((..........))))))).(((((..((....))..))))))))..-(((.((((....(((.---(((---......)).)....-))).--..)))).))). ( -33.20) >DroWil_CAF1 94398 88 - 1 CUAGCCACAAAUUAAACGAAUUGUGGCACAGAA-CUGGCAACAUUUGUUGCCACA-UUGACCAGA---UGUA---GCA---ACAAC------------UG--CAGCAGGAAAC ...(((((((.((....)).)))))))......-((.((..((.(((((((.(((-((.....))---))).---)))---)))).------------))--..)).)).... ( -27.90) >DroYak_CAF1 89048 93 - 1 UUUGCCGCAAAUGAAACGAAUUGUGGCACUGGAUUUGGCAACAUUUGUUGCCACAGAUGACCGGCACUUGGA---GCA---UCAUC------------AU--CACCAGGAUGC ..((((((((..........))))))))((((...(((((((....)))))))..(((((...((.......---)).---)))))------------..--..))))..... ( -33.00) >DroAna_CAF1 82014 106 - 1 GUGGCCGCAAAUUAAACGAAUUGUGGCUCGGCAUUUGGCAACAUUUGUUGCCACA-CUGACCAG---UUGGGAAAGUA---UCAGCAUCAUCAGCCGUGCCGGGUCAGGAAAC ((..((((((.((....)).))))((((((((((.(((((((....)))))))..-((((...(---((((.......---)))))....))))..)))))))))).))..)) ( -36.80) >consensus UUGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA_CUGACCGGCACUUGGA___GCA___GCAGCAU_AUCAUC__CAU__CACCAGGAAGC ...(((((((..........))))))).(((....(((((((....)))))))...)))...................................................... (-17.69 = -18.33 + 0.64)

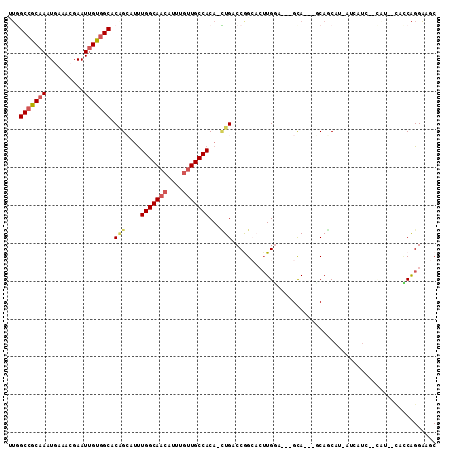
| Location | 23,288,706 – 23,288,798 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
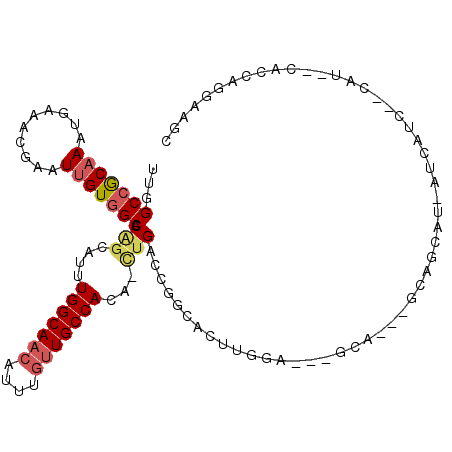
>3R_DroMel_CAF1 23288706 92 + 27905053 -UC----CAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAGGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAACGGGAACGG------GAACGGGUCG--AG -..----...(((((((((..-..(((((((....)))))))..))))).).))).(((((((((.(((.((.....)).)))..)------))))))))..--.. ( -32.00) >DroSec_CAF1 86325 102 + 1 CUGCUGCUCAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAA-GCUGUGCCACAAUUCGUUUCAUUUGCGGCCAACGGGAACGGGAACGAGAACGAGUCG--AG ...(.((((......((((((-(.(((((((....)))))))..-)))).)))....((((((((.(((.((.....)).)))..))))))))...)))).)--.. ( -35.70) >DroSim_CAF1 89474 86 + 1 CUC----CAAGUGCCGGUCAG-UGUGGCAC--AAUGUUGCCAAAGGCUGUGC-AC-AUUCGUUUCAUUUGCGGCCAC--GGAACG-------GAACGGGUCG--AG (((----...(((((((((..-..(((((.--.....)))))..))))).))-))-(((((((((..((.((....)--).)).)-------)))))))).)--)) ( -30.40) >DroEre_CAF1 91791 82 + 1 -UG----CAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCCACGG------------------GUCGCAGG -.(----(....)).((((((-(.(((((((....)))))))...)))).)))..............((((((((....)------------------))))))). ( -33.40) >DroWil_CAF1 94419 89 + 1 -UA----CA---UCUGGUCAA-UGUGGCAACAAAUGUUGCCAG-UUCUGUGCCACAAUUCGUUUAAUUUGUGGCUAGAG--CACA---UUGAGAAGUGGACA--AG -..----((---.((..((((-(((((((((....)))))).(-(((((.(((((((..........))))))))))))--))))---))))..))))....--.. ( -37.30) >DroYak_CAF1 89069 88 + 1 -UC----CAAGUGCCGGUCAUCUGUGGCAACAAAUGUUGCCAAAUCCAGUGCCACAAUUCGUUUCAUUUGCGGCAAACGG-------------CAGGAGUCGCAGG -((----(...(((((.....((((((((((....)))))))....)))((((.(((..........))).))))..)))-------------)))))........ ( -29.00) >consensus _UC____CAAGUGCCGGUCAG_UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAACGG_AACG_______GAACGGGUCG__AG ............(((((((((...(((((((....)))))))....))).))).......((.......)))))................................ (-15.66 = -16.55 + 0.89)

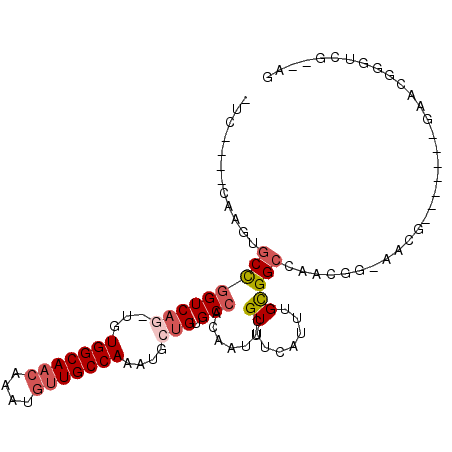

| Location | 23,288,706 – 23,288,798 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.63 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23288706 92 - 27905053 CU--CGACCCGUUC------CCGUUCCCGUUGGCCGCAAAUGAAACGAAUUGUGGCACAGCCUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUG----GA- ..--....(((...------(((.((..(((((((((((..........))))))).))))...(((((((....)))))))..-..)).)))....))----).- ( -30.20) >DroSec_CAF1 86325 102 - 1 CU--CGACUCGUUCUCGUUCCCGUUCCCGUUGGCCGCAAAUGAAACGAAUUGUGGCACAGC-UUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUGAGCAGCAG ..--......(((((((...(((.((..(((((((((((..........))))))).))))-..(((((((....)))))))..-..)).)))...)))).))).. ( -34.10) >DroSim_CAF1 89474 86 - 1 CU--CGACCCGUUC-------CGUUCC--GUGGCCGCAAAUGAAACGAAU-GU-GCACAGCCUUUGGCAACAUU--GUGCCACA-CUGACCGGCACUUG----GAG ..--........((-------((((((--(.(((.(((.((.......))-.)-))...)))..))).)))...--(((((...-......)))))..)----)). ( -21.40) >DroEre_CAF1 91791 82 - 1 CCUGCGAC------------------CCGUGGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUG----CA- ((..((..------------------.))..))..((((.((...((.(.(((((((..(((..((....))..))))))))))-.)...)).)).)))----).- ( -28.30) >DroWil_CAF1 94419 89 - 1 CU--UGUCCACUUCUCAA---UGUG--CUCUAGCCACAAAUUAAACGAAUUGUGGCACAGAA-CUGGCAACAUUUGUUGCCACA-UUGACCAGA---UG----UA- ..--......((..((((---(((.--.....(((((((.((....)).)))))))......-..((((((....)))))))))-))))..)).---..----..- ( -27.90) >DroYak_CAF1 89069 88 - 1 CCUGCGACUCCUG-------------CCGUUUGCCGCAAAUGAAACGAAUUGUGGCACUGGAUUUGGCAACAUUUGUUGCCACAGAUGACCGGCACUUG----GA- ........(((((-------------(((..((((((((..........))))))))(((....(((((((....)))))))))).....)))))...)----))- ( -30.70) >consensus CU__CGACCCGUUC_______CGUU_CCGUUGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA_CUGACCGGCACUUG____GA_ ....((..........................(((((((..........))))))).(((....(((((((....)))))))...)))..)).............. (-18.39 = -18.87 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:52 2006