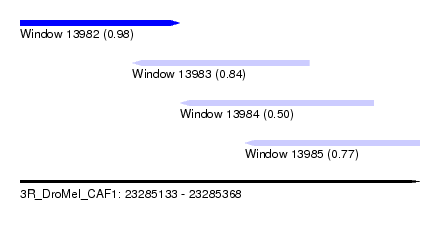
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,285,133 – 23,285,368 |
| Length | 235 |
| Max. P | 0.977681 |

| Location | 23,285,133 – 23,285,227 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -13.97 |
| Energy contribution | -15.63 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
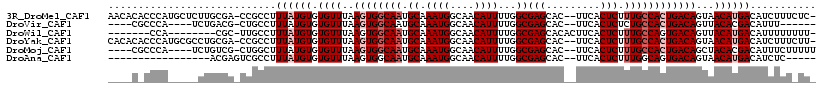
>3R_DroMel_CAF1 23285133 94 + 27905053 CCGGGUUGGCUUUU-GCAAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACUAAAAGUUGGCAACAAAAAA-AAAAA--AAAAAAAGAAGG- ....((..((((((-((.(((.(((((((((....)))))))))......))).)))...)))))..)).........-.....--............- ( -21.20) >DroGri_CAF1 96514 83 + 1 ACAAG-UUGCCACU-G--CAAGUGGA-UGAAAAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUUCCAAAAAAAAAGGGAAAUAA-----------A ...((-(((((((.-.--...)))((-((((....))))))))))))................((((((..........))))))..-----------. ( -14.50) >DroSec_CAF1 82794 96 + 1 CCGGGUUGGCUUUU-GCAAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUGGCAACAAAAAA-AAAAAUAAAAAAAGGAAGG- ....((..((((((-((.(((.(((((((((....)))))))))......))).)))...)))))..)).........-...................- ( -21.20) >DroSim_CAF1 85934 96 + 1 CCGGGUUGGCUUUU-GCAAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUGGCAACAAAAAA-AAAAAUAAAAAAAGGAAGG- ....((..((((((-((.(((.(((((((((....)))))))))......))).)))...)))))..)).........-...................- ( -21.20) >DroEre_CAF1 88340 74 + 1 ------UGGCUUUU-GCGAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUGGCAACAA-------------AAA----AAGG- ------(((((...-((....))((((((((....))))))))..........))).)).....(((....)))-------------...----....- ( -17.10) >DroYak_CAF1 85399 93 + 1 CCGGGUUGGCUUUUGGCGAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUGGCAACAGAGAA-AAAAAAAGAAA----AAGG- ....((..((((((.((.(((.(((((((((....)))))))))......))).))...))))))..)).........-...........----....- ( -23.00) >consensus CCGGGUUGGCUUUU_GCAAAAGUGCAGUGAACAUUUUCAUUGCAAUUAAAUUUAGCACAAAAAGUUGGCAACAAAAAA_AAAAAUAAAAA____AAGG_ ....((..((((((.((.(((.(((((((((....)))))))))......))).))...))))))..)).............................. (-13.97 = -15.63 + 1.67)


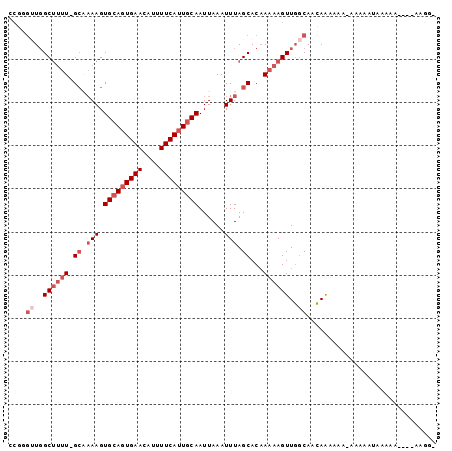
| Location | 23,285,199 – 23,285,303 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23285199 104 - 27905053 UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUUCUC--CCUUCU---UUUUUU--UU--UUU-U--UUUUUGUUGC ..((((((((.((.((((....))))...))(((...--....))).))))))))....((((((.............--......---......--..--...-.--....)))))) ( -21.85) >DroGri_CAF1 96576 98 - 1 UAAGUGGCAAUGCAAACGGCAACAUUUUGGCGAGCAC--UUUUCUCUGUGCCACUGACAGUUACACGACAUUCGUUUUUU--------------UUAUU--UCCCU--UUUUUUUUGG ...(((((..(((.....))).....(..(.(.((((--........))))).)..)..)))))(((.....))).....--------------.....--.....--.......... ( -16.00) >DroEre_CAF1 88400 90 - 1 UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUUCUC--CCUU-------UUU-----------------UUGUUGC ..((((((((.((.((((....))))...))(((...--....))).))))))))....((((((.............--....-------...-----------------.)))))) ( -22.40) >DroWil_CAF1 90815 102 - 1 UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACACUUCACUCUUUGCCAGUGACAGUUACAUGACAUUUUUUUU-------------UUCUUGUU--UUU-UCGCUUUUUUGGC .(((((((..(((.((((....))))...))).)).)))))........(((((.((.(((.....((((........-------------....))))--...-..))))).))))) ( -20.20) >DroMoj_CAF1 98240 113 - 1 UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGCUACACGACAUUUCUUUUUUUCUGUGUGAUUUU-UUUUUUCCCCCU--UUUUUUUUGC ..((((((((.((.((((....))))...))(((...--....))).))))))))......((((((...............)))))).....-............--.......... ( -22.56) >DroAna_CAF1 78572 87 - 1 UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGGCAGUGACAGUAACAUGACAUCUC-------CUU-------U---UAUU------------UUUUGGC ...(((((..(((.((((....))))...))).))((--(((((((....).))))).)))..)))........-------...-------.---....------------....... ( -14.50) >consensus UAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC__UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUUUUU___CUU_______UU_UU_UU__U___U__UUUUUUUUGC ..((((((((.((.((((....))))...))(((.........))).))))))))............................................................... (-14.90 = -15.73 + 0.83)

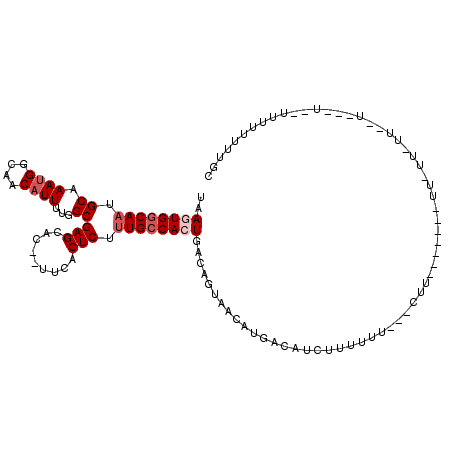
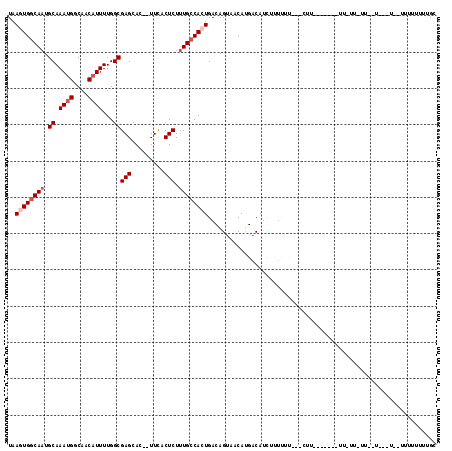
| Location | 23,285,227 – 23,285,341 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23285227 114 - 27905053 AACACACCCAUGCUCUUGCGA-CCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUUCUC- ..........(((....))).-......((((((.((((..((((((((.((.((((....))))...))(((...--....))).))))))))))))...))))))..........- ( -27.70) >DroVir_CAF1 94162 101 - 1 ----CGCCCA----UCUGACG-CUGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUCUGCCACUGACAGUUACACGACAUUU------ ----......----.......-.........((((((....(((((((..((.((((....))))...))(((...--....)))..)))))))......))))))......------ ( -24.80) >DroWil_CAF1 90839 101 - 1 -------CCA--------CGC-UUGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACACUUCACUCUUUGCCAGUGACAGUUACAUGACAUUUUUUUU- -------...--------.((-(((((...((((.((((.....)))).(((.....)))))))...)))))))..((((((((.......))))).))).................- ( -23.90) >DroYak_CAF1 85492 114 - 1 CACACACCCAUGCGCCUGCGA-CCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUUCUU- ((((((.....(((.......-.))).....))))))....((((((((.((.((((....))))...))(((...--....))).)))))))).......................- ( -28.70) >DroMoj_CAF1 98276 107 - 1 ----CGCCCA----UCUGUCG-CUGGCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGCCACUGACAGCUACACGACAUUUCUUUUU ----.(((..----.......-..)))....((((((((..((((((((.((.((((....))))...))(((...--....))).))))))))...))).)))))............ ( -28.90) >DroAna_CAF1 78587 94 - 1 -----------------ACGAGUCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC--UUCACUCUUUGGCAGUGACAGUAACAUGACAUCUC----- -----------------..((((((((...((((.((((.....)))).(((.....)))))))...)))))..((--(((((((....).))))).)))..........)))----- ( -21.10) >consensus ____C_CCCA_____CUGCGA_CCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCAC__UUCACUCUUUGCCACUGACAGUAACAUGACAUCUUU_UU_ ............................((((((.((((..((((((((.((.((((....))))...))(((.........))).))))))))))))...))))))........... (-20.32 = -21.32 + 1.00)

| Location | 23,285,265 – 23,285,368 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23285265 103 - 27905053 CCCAGCU-------------ACCACCACCUUUGCCGCACCAACACACCCAUGCUCUUGCGACCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACU ....(((-------------.(((......(((((((...(((((((....((....))(.....).....)))))))...)))))))...(((((....))))))))..)))... ( -28.20) >DroSec_CAF1 82928 108 - 1 CCCAGCUACU-------CCCACCACCACC-UUGCUGCACCAACACACCCAUGCUCCUGCGACCGCCCUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACU ........((-------((((........-((((..(...(((((((....((....))(......)....)))))))...)..))))...(((((....)))))))).))).... ( -25.40) >DroSim_CAF1 86068 115 - 1 CCCAGCUACUACCUCCACCCACCCCCACC-UUGCCGCACCAACACACCCAUGCUCCUGCGACCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACU ....(((......................-(((((((...(((((((....((....))(.....).....)))))))...)))))))(((.((((....))))...))))))... ( -27.80) >DroEre_CAF1 88452 101 - 1 CCCAACU-------------GCCC-CAAC-UUGCGGCAGCCACACACCCAUGCUCCUGCGACCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACU ......(-------------((((-(((.-.((((...(((((((((.((((.....((....))...)))).))))....)))))..))))((((....)))))))).).))).. ( -28.20) >consensus CCCAGCU_____________ACCACCACC_UUGCCGCACCAACACACCCAUGCUCCUGCGACCGCCUUUAUGUGUGUUUAAGUGGCAAUGCAAAUGGCAACAUUUUGGCGAGCACU ....(((.......................(((((((...(((((((....((....))(.....).....)))))))...)))))))(((.((((....))))...))))))... (-23.05 = -23.68 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:46 2006