| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,270,377 – 23,270,494 |
| Length | 117 |
| Max. P | 0.976471 |

| Location | 23,270,377 – 23,270,494 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
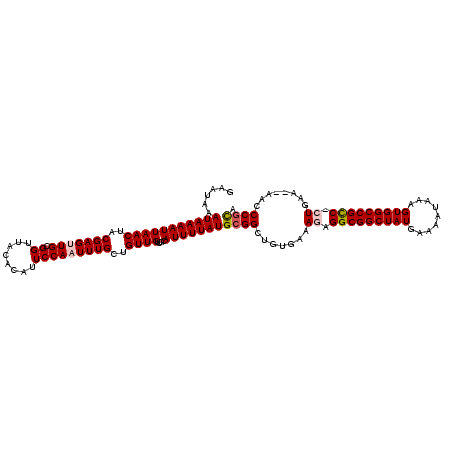
>3R_DroMel_CAF1 23270377 117 - 27905053 AAAUAAAUAAAAUUAACUACGAGUUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAGAGGCGGCUAUGAAAAUAAAGUGGCCGUC-CUGUA--AACCCGCA ......(((((((((((..(((((((.((........)))))))))..))))....)))))))((((..((...((.(((((((((.........)))))))))-))...--.)))))). ( -32.00) >DroVir_CAF1 75218 117 - 1 GAAUAAAUAAAAUUAACUACGAGCUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAAAGGCGGCUAUGAAAAUAAAGUGGCCGCC-AUGAA--AACCCGUA .................((((((((((((.........))((..(((.......)))..))..))))))........(((((((((.........)))))))))-.....--....)))) ( -31.80) >DroPse_CAF1 71760 117 - 1 GAAUAAAUAAAAUUAACUACGAGUUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAGAGGCGGCUAUGAAAAUAAAGUGGCCGCCCCUGAA--AGCCCGC- ......(((((((((((..(((((((.((........)))))))))..))))....)))))))((((((.....((.(((((((((.........))))))))).))...--)).))))- ( -34.90) >DroGri_CAF1 77487 117 - 1 GUAUAAAUAAAAUUAACUACGAGCUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAAAGGCGGCUAUGAAAAUAAAGUGGCCGCC-AUGAA--AGCCCGCA ......(((((((((((.((.(((...((.........)).....))))).)))).)))))))((((((........(((((((((.........)))))))))-.....--)).)))). ( -32.32) >DroWil_CAF1 74336 120 - 1 GAAUAAAUAAAAUUAACUACGAGUUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAGAGGCGGCUAUGAAAAUAAAGUGGCCGUCUCUGAAAAAACCCUCA ....................(((....((((...........((..(....(((((((...))))))).)..))((((((((((((.........)))))))))))).....))))))). ( -32.60) >DroPer_CAF1 72022 117 - 1 GAAUAAAUAAAAUUAACUACGAGUUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAGAGGCGGCUAUGAAAAUAAAGUGGCCGCCCCUGAA--AGCCCGC- ......(((((((((((..(((((((.((........)))))))))..))))....)))))))((((((.....((.(((((((((.........))))))))).))...--)).))))- ( -34.90) >consensus GAAUAAAUAAAAUUAACUACGAGUUGCGGUUACACAUUCCAAUUUGCUGUUGUUGCAUUUUAUGCGGCUGUGAAAGAGGCGGCUAUGAAAAUAAAGUGGCCGCC_CUGAA__AACCCGCA ......(((((((((((..(((((((.((........)))))))))..))))....)))))))((((.......((.(((((((((.........))))))))).))........)))). (-29.20 = -29.68 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:41 2006