
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,255,989 – 23,256,114 |
| Length | 125 |
| Max. P | 0.993314 |

| Location | 23,255,989 – 23,256,081 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23255989 92 + 27905053 AAUUUAU---GUUACUAAGUUACAUAUUUUACAUUUGUGAGGUCUCUUUUUUCACAGUGCUUUUAUCUGCUGAUUAUGUGCUAGAUGGUAUGCAU ((..(((---((.((...)).)))))..)).....(((((((.......)))))))((((..((((((((.........)).))))))...)))) ( -17.00) >DroSec_CAF1 53464 76 + 1 GACUUAA---GUUACUGAGUUACUCA--------------GAAGC--UUUUUUUCAGUGCAUUUAUCUGCUUAUUAUGUGCUAGAUGGUAUGCAU ((...((---(((.(((((...))))--------------).)))--))....)).((((((((((((((.........)).)))))).)))))) ( -20.50) >DroSim_CAF1 53393 81 + 1 AACAUUACAUUUUACUGAGUUACUCA--------------GAUACAUUUUUUUUCAGUGCAUUUAUCUGCUUAUUAUGUGCUAGAUGGUAUGCAU ..............(((((...))))--------------)...............((((((((((((((.........)).)))))).)))))) ( -16.40) >consensus AACUUAA___GUUACUGAGUUACUCA______________GAUAC_UUUUUUUUCAGUGCAUUUAUCUGCUUAUUAUGUGCUAGAUGGUAUGCAU ........................................................((((((((((((((.........)).)))))).)))))) (-10.40 = -10.73 + 0.33)



| Location | 23,256,016 – 23,256,114 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -12.30 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23256016 98 + 27905053 ACAUUUGUGAGGUCU--CUUUUUUCACAGUGCUUUUAUCUGCU--GAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUC .(((((((((((...--....))))))((..(....(((....--)))...)..)))))))((((((((.......))))))))....(((....))).... ( -23.00) >DroSec_CAF1 53487 86 + 1 ----------GAAGC----UUUUUUUCAGUGCAUUUAUCUGCU--UAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUC ----------..(((----.........((((((((((((((.--........)).)))))).))))))(((((....))))).....(((....)))))). ( -18.50) >DroSim_CAF1 53419 88 + 1 ----------GAUAC--AUUUUUUUUCAGUGCAUUUAUCUGCU--UAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUC ----------.((((--((........(((((((((((((((.--........)).)))))).)))))))))))))............(((....))).... ( -17.80) >DroEre_CAF1 57803 82 + 1 -----------------GAUUUUUUGCCGUGCAC-UAUCUGCU--UAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUC -----------------(((...(((.((((.((-(((((((.--........)).)))))))((((((.......))))))...)))).)))..))).... ( -18.60) >DroYak_CAF1 53314 81 + 1 -----------------UAUUUUUUGCAGUGCAUUUAUCUGCU--UAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGC-- -----------------........(((((.(((.(((((((.--........)).)))))((((((((.......))))))))..)))...)))))...-- ( -18.00) >DroAna_CAF1 47391 88 + 1 ----------GAAGCCUUUUUUCUUCCUGU----CUAUACUCUUAUAUAAUGUGCUGGAUAUUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUU ----------.((((............(((----((((((...........))).)))))).(((((((.......))))))).....(((....))))))) ( -15.80) >consensus __________GA__C__UUUUUUUUGCAGUGCAUUUAUCUGCU__UAUUAUGUGCUAGAUGGUAUGCAUUAUGUAUGUGCAUAUUUAUGACAACUGUCGCUC ...................................(((((((...........)).)))))((((((((.......))))))))....(((....))).... (-12.30 = -12.38 + 0.09)


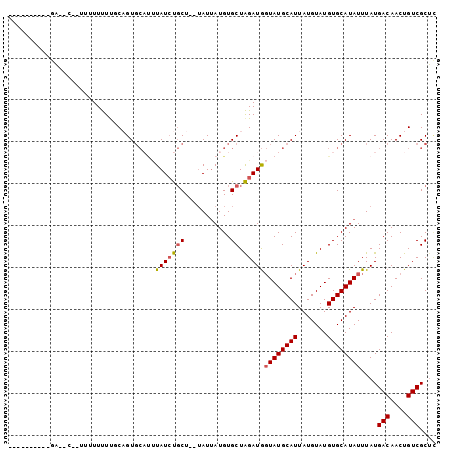
| Location | 23,256,016 – 23,256,114 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -13.50 |
| Consensus MFE | -9.67 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23256016 98 - 27905053 GAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUC--AGCAGAUAAAAGCACUGUGAAAAAAG--AGACCUCACAAAUGU ..(((((....))).....((((((.........))))))..((((.((........--.))))))....))..(((((......--.....)))))..... ( -15.00) >DroSec_CAF1 53487 86 - 1 GAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUA--AGCAGAUAAAUGCACUGAAAAAAA----GCUUC---------- (((((((....))).....((((......)))).(((((...((((.((........--.))))))..)))))..........----)))).---------- ( -14.60) >DroSim_CAF1 53419 88 - 1 GAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUA--AGCAGAUAAAUGCACUGAAAAAAAAU--GUAUC---------- ..(((((....))).....((((((.........)))))).......))........--.(((......))).............--.....---------- ( -12.50) >DroEre_CAF1 57803 82 - 1 GAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUA--AGCAGAUA-GUGCACGGCAAAAAAUC----------------- ........((((((((...((((((.........))))))..((((.((........--.)))))).-))).)))))........----------------- ( -13.40) >DroYak_CAF1 53314 81 - 1 --GCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUA--AGCAGAUAAAUGCACUGCAAAAAAUA----------------- --(((((....))).....((((((.........)))))).......))........--.((((.........))))........----------------- ( -13.40) >DroAna_CAF1 47391 88 - 1 AAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUAAUAUCCAGCACAUUAUAUAAGAGUAUAG----ACAGGAAGAAAAAAGGCUUC---------- (((((((....))).....((((((.........))))))...(((......((((((....))))))----...))).........)))).---------- ( -12.10) >consensus GAGCGACAGUUGUCAUAAAUAUGCACAUACAUAAUGCAUACCAUCUAGCACAUAAUA__AGCAGAUAAAUGCACUGAAAAAAAAA__G__UC__________ ..(((((....))).....((((((.........)))))).......))...........(((......))).............................. ( -9.67 = -10.33 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:38 2006