
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,243,690 – 23,243,822 |
| Length | 132 |
| Max. P | 0.999659 |

| Location | 23,243,690 – 23,243,791 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -22.04 |
| Energy contribution | -24.85 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23243690 101 + 27905053 UUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAAAAU----UUGAGGGGAAACCAGCAGCUCGUUACAGCUGCA-AAUAAC (((((((((.....))))..)))))..((((.(..((((((((.....((((((((((....----)))))))))).......))))))))..))))).-...... ( -36.10) >DroSec_CAF1 41387 101 + 1 UUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAU----UUGAGGGGAAACCAGCAGCUCGCUACAGCUGCA-AAUAAC ..(((((.((...((.....))...)).)))))...............((((((((((....----))))))))))....((((((......)))))).-...... ( -33.20) >DroSim_CAF1 41277 101 + 1 UUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAU----UUGAGGGGAAAGCAGCAGCUCGUUACAGCUGCA-AAUAAC (((((((((.....))))..)))))..((((.(..((((((((..(..((((((((((....----))))))))))..)....))))))))..))))).-...... ( -37.70) >DroEre_CAF1 46192 99 + 1 UUGUCUG-GCAAAUCCUAAUGACGAGUGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAU----UCGAGGG-AAACCAGCAGCUCGUUACCGCUGCA-AAUAAC (((((.(-(.....))....))))).(((((....((((((((.....(((((((.......----..)))))-)).......))))))))...)))))-...... ( -29.10) >DroYak_CAF1 41012 101 + 1 UUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGAGUUCCCUUCAAUAAU----UUGAGGGUAAACCAGCAGCUCGUUACUGCUGCA-AAUAAC (((((((((.....))))..)))))..((((.(..((((((((....(((.(((((((....----)))))))..))).....))))))))..))))).-...... ( -31.50) >DroAna_CAF1 35672 85 + 1 ---------CAAAUACU----------CUAACCCGUAACUAGGACCU-GUCCCACCAAUAAUAAUUUUUUGGGGAAACCA-CUGCUCGUUAUUGCAACGGAGUAAC ---------....((((----------((............(((...-.)))...(((((((.........((....)).-......)))))))....)))))).. ( -17.09) >consensus UUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAU____UUGAGGGGAAACCAGCAGCUCGUUACAGCUGCA_AAUAAC (((((((((.....))))..)))))..((((.(..((((((((.....((((((((((........)))))))))).......))))))))..)))))........ (-22.04 = -24.85 + 2.81)



| Location | 23,243,690 – 23,243,791 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23243690 101 - 27905053 GUUAUU-UGCAGCUGUAACGAGCUGCUGGUUUCCCCUCAA----AUUUUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAA (((...-.(((((((((((((((....(..(((((.((((----....)))).)))))..)..))))))))))).))))((..(((.....)))..))...))).. ( -40.20) >DroSec_CAF1 41387 101 - 1 GUUAUU-UGCAGCUGUAGCGAGCUGCUGGUUUCCCCUCAA----AUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAA (((...-.(((((((((((((((....(..(((((.((((----....)))).)))))..)..))))))))))).))))((..(((.....)))..))...))).. ( -40.20) >DroSim_CAF1 41277 101 - 1 GUUAUU-UGCAGCUGUAACGAGCUGCUGCUUUCCCCUCAA----AUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAA (((...-.(((((((((((((((....((((.(((.((((----....)))).)))))))...))))))))))).))))((..(((.....)))..))...))).. ( -37.80) >DroEre_CAF1 46192 99 - 1 GUUAUU-UGCAGCGGUAACGAGCUGCUGGUUU-CCCUCGA----AUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCACUCGUCAUUAGGAUUUGC-CAGACAA ......-((((((.(((((((((....(..((-(((((((----....))).))))))..)..))))))))).).)))))...(((....((.....)-).))).. ( -37.40) >DroYak_CAF1 41012 101 - 1 GUUAUU-UGCAGCAGUAACGAGCUGCUGGUUUACCCUCAA----AUUAUUGAAGGGAACUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAA ......-.......(((((((((....((((..(((((((----....))).))))))))...))))))))).(((((.((..(((.....)))..)).))))).. ( -32.60) >DroAna_CAF1 35672 85 - 1 GUUACUCCGUUGCAAUAACGAGCAG-UGGUUUCCCCAAAAAAUUAUUAUUGGUGGGAC-AGGUCCUAGUUACGGGUUAG----------AGUAUUUG--------- .(((..((((..((((((..(....-(((.....))).....)..)))))).(((((.-...)))))...))))..)))----------........--------- ( -17.40) >consensus GUUAUU_UGCAGCUGUAACGAGCUGCUGGUUUCCCCUCAA____AUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAA ........(((((..((((((((....(..(((((.((((........)))).)))))..)..))))))))..).))))........................... (-24.83 = -25.80 + 0.97)


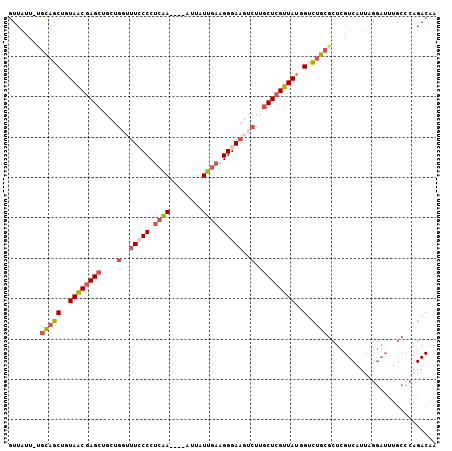
| Location | 23,243,730 – 23,243,822 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23243730 92 + 27905053 AGCAAGACUUCCCUUCAAAAAUUUGAGGGGAAACCAGCAGCUCGUUACAGCUGCAAAUAACUGAGUUUGAUGCUGACUUUCUUUUCAGUUUU--------- ........((((((((((....))))))))))....((((((......))))))....(((((((...((.........))..)))))))..--------- ( -28.40) >DroSec_CAF1 41427 92 + 1 AGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAACCAGCAGCUCGCUACAGCUGCAAAUAACUGAGUUUGAUGCUGACUUUCUUUUCAGUUUU--------- ........((((((((((....))))))))))....((((((......))))))....(((((((...((.........))..)))))))..--------- ( -29.20) >DroSim_CAF1 41317 92 + 1 AGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAAGCAGCAGCUCGUUACAGCUGCAAAUAACUGAGUUUGAUGCUGACUUUCUUUUCAGUUUU--------- .....(..((((((((((....))))))))))..).((((((......))))))....(((((((...((.........))..)))))))..--------- ( -30.10) >DroEre_CAF1 46231 100 + 1 AGCAAGACUUCCCUUCAAUAAUUCGAGGG-AAACCAGCAGCUCGUUACCGCUGCAAAUAACUGAGUUUGCUGCUGACUUUCUUUUCGGUUUUCGGUUUUCG ((((((..(((((((.........)))))-))....(((((........)))))...........))))))(((((........)))))............ ( -24.10) >DroYak_CAF1 41052 101 + 1 AGCAAGAGUUCCCUUCAAUAAUUUGAGGGUAAACCAGCAGCUCGUUACUGCUGCAAAUAACUGAGUUUGAUGCUGACUUUCUUUUCAGUUUUUCGUUUUCA ......((((.(((((((....))))))).......(((((........)))))....))))(((..(((.(((((........)))))...)))..))). ( -27.00) >consensus AGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAACCAGCAGCUCGUUACAGCUGCAAAUAACUGAGUUUGAUGCUGACUUUCUUUUCAGUUUU_________ .....(..((((((((((....))))))))))..).(((((........)))))....(((((((...((.........))..)))))))........... (-24.22 = -24.86 + 0.64)


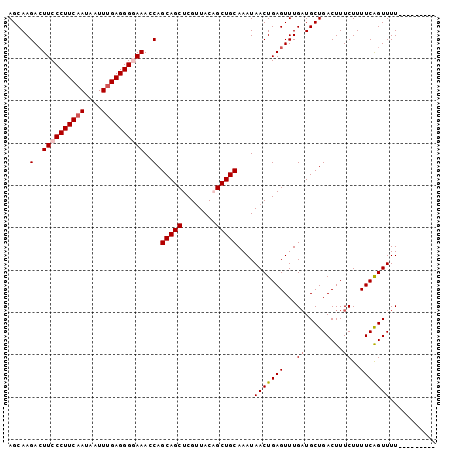
| Location | 23,243,730 – 23,243,822 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23243730 92 - 27905053 ---------AAAACUGAAAAGAAAGUCAGCAUCAAACUCAGUUAUUUGCAGCUGUAACGAGCUGCUGGUUUCCCCUCAAAUUUUUGAAGGGAAGUCUUGCU ---------....((....))......((((................((((((......)))))).(..(((((.((((....)))).)))))..).)))) ( -25.90) >DroSec_CAF1 41427 92 - 1 ---------AAAACUGAAAAGAAAGUCAGCAUCAAACUCAGUUAUUUGCAGCUGUAGCGAGCUGCUGGUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCU ---------....((....))......((((................((((((......)))))).(..(((((.((((....)))).)))))..).)))) ( -25.90) >DroSim_CAF1 41317 92 - 1 ---------AAAACUGAAAAGAAAGUCAGCAUCAAACUCAGUUAUUUGCAGCUGUAACGAGCUGCUGCUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCU ---------....((....))......((((................((((((......))))))...((((((.((((....)))).))))))...)))) ( -23.40) >DroEre_CAF1 46231 100 - 1 CGAAAACCGAAAACCGAAAAGAAAGUCAGCAGCAAACUCAGUUAUUUGCAGCGGUAACGAGCUGCUGGUUU-CCCUCGAAUUAUUGAAGGGAAGUCUUGCU ..................((((...((((((((....((.((((((......))))))))))))))))(((-(((((((....))).)))))))))))... ( -25.60) >DroYak_CAF1 41052 101 - 1 UGAAAACGAAAAACUGAAAAGAAAGUCAGCAUCAAACUCAGUUAUUUGCAGCAGUAACGAGCUGCUGGUUUACCCUCAAAUUAUUGAAGGGAACUCUUGCU ......((((.((((((....................)))))).))))(((((((.....))))))).....(((((((....))).)))).......... ( -23.05) >consensus _________AAAACUGAAAAGAAAGUCAGCAUCAAACUCAGUUAUUUGCAGCUGUAACGAGCUGCUGGUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCU ...........................((((................(((((........))))).(..(((((.((((....)))).)))))..).)))) (-19.96 = -20.20 + 0.24)

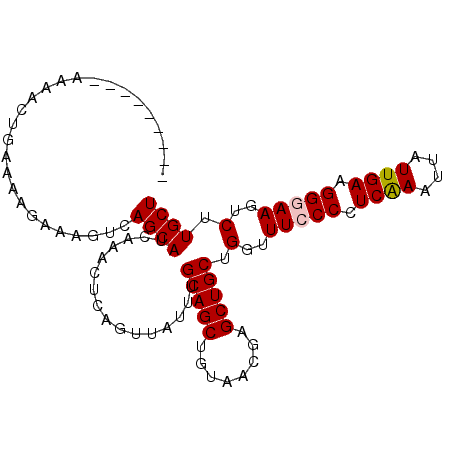
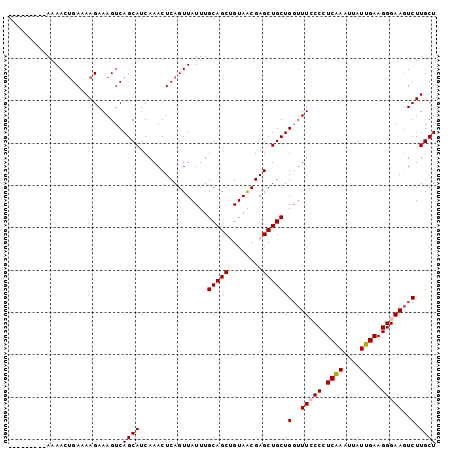
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:34 2006