
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,210,923 – 23,211,017 |
| Length | 94 |
| Max. P | 0.961593 |

| Location | 23,210,923 – 23,211,017 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23210923 94 + 27905053 ----CCAUUUUCGUUGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCAGAAUUGAAACAUUGCCAGGACCAGCUCCUUAC ----........(((((..(((..((((((((((.......)))((((((.......))))))........)))))...))..))))))))....... ( -27.60) >DroPse_CAF1 9075 89 + 1 U---GCUCUUUCCUUGGCAGAGCAACAUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCUCCGGAAUUGAAACAUUGCCAACAUCA------UGC .---((.......((((((((((.........))))((.((.((((((((.......))))).))).)).))......)))))).....------.)) ( -26.82) >DroSec_CAF1 8539 90 + 1 ----CCAUUUUCGUUGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCCGAAUUGAAACAUUGCCAG----GACUCCAUAC ----......((.((((((.....(((....)))((.(((((.(((((((.......))))))).))))).)).....))))))----))........ ( -27.00) >DroWil_CAF1 8261 83 + 1 ----CGAU----GGUGGCAGG-CAGCAUUUCCGCUCCAUUUGCGGGCAGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAACAUCA------UGC ----.(((----(.((((((.-...((.((((((.......))((((.((.......)).)))).)))).)).....)))))).)))).------... ( -27.80) >DroAna_CAF1 2810 85 + 1 UGCCACAUUUUUGUUGGCAGGGCAGCAUUUCCGCUCCAUUUGCGGGCCGUCAAAGGAAAGGCCUCGGAAUUGAAACAUUGCCA-------------AC (((((((....)).))))).(((((((.((((((.......))(((((.((....))..))))).)))).)).....))))).-------------.. ( -27.90) >DroPer_CAF1 9121 89 + 1 U---GCUCUUUCCUUGGCAGAGAAACAUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCUCCGGAAUUGAAACAUUGCCAACAUCA------UGC .---((.......(((((((.(...((.((((((.......))(((((((.......))))).)))))).))...).))))))).....------.)) ( -26.22) >consensus ____CCAUUUUCGUUGGCAGAGAAGCAUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAACAUCA______UAC ..............((((((.....((.((((((.......))(((((((.......))))))).)))).)).....))))))............... (-22.22 = -22.55 + 0.33)



| Location | 23,210,923 – 23,211,017 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23210923 94 - 27905053 GUAAGGAGCUGGUCCUGGCAAUGUUUCAAUUCUGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCAACGAAAAUGG---- ....((.((((.((..((((((((((((....))))).)))))))..))))))))(((..(.(((((....))))).).)))............---- ( -33.80) >DroPse_CAF1 9075 89 - 1 GCA------UGAUGUUGGCAAUGUUUCAAUUCCGGAGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAAUGUUGCUCUGCCAAGGAAAGAGC---A ((.------(..(.((((((..((..((.((((((.((((((.....))))))))((.......)))))).))..))..)))))).)..)..))---. ( -32.90) >DroSec_CAF1 8539 90 - 1 GUAUGGAGUC----CUGGCAAUGUUUCAAUUCGGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCAACGAAAAUGG---- ((.(((.(((----(.(((...((((((.(((((..((((((.....))))))))).)).))))))(....)))).)))).)))))........---- ( -34.10) >DroWil_CAF1 8261 83 - 1 GCA------UGAUGUUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACUGCCCGCAAAUGGAGCGGAAAUGCUG-CCUGCCACC----AUCG---- ...------.((((.(((((..((..((.(((((...(((((((...........)).)))))..))))).))..)-).))))).)----))).---- ( -28.30) >DroAna_CAF1 2810 85 - 1 GU-------------UGGCAAUGUUUCAAUUCCGAGGCCUUUCCUUUGACGGCCCGCAAAUGGAGCGGAAAUGCUGCCCUGCCAACAAAAAUGUGGCA ((-------------(((((..((..((.(((((.((((((......)).)))))((.......)))))).))..))..)))))))............ ( -27.10) >DroPer_CAF1 9121 89 - 1 GCA------UGAUGUUGGCAAUGUUUCAAUUCCGGAGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAAUGUUUCUCUGCCAAGGAAAGAGC---A ((.------(..(.((((((..((((((.((.(((.((((((.....))))))))).)).))))))((((....)))).)))))).)..)..))---. ( -30.30) >consensus GCA______UGAUGUUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAAUGCUGCCCUGCCAACGAAAAUGG____ ...............(((((....((((.((.((.(((((((.....))))))))).)).))))(((....))).....))))).............. (-21.00 = -21.28 + 0.28)

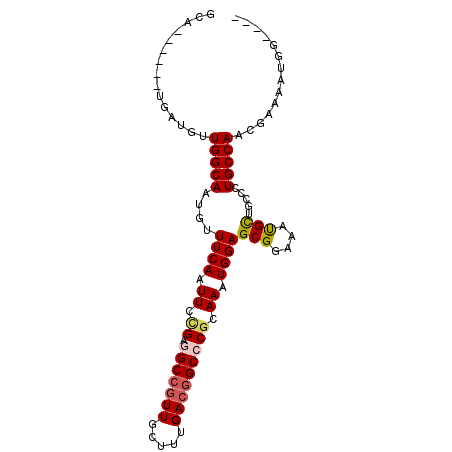
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:23 2006