
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,206,173 – 23,206,273 |
| Length | 100 |
| Max. P | 0.993692 |

| Location | 23,206,173 – 23,206,273 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
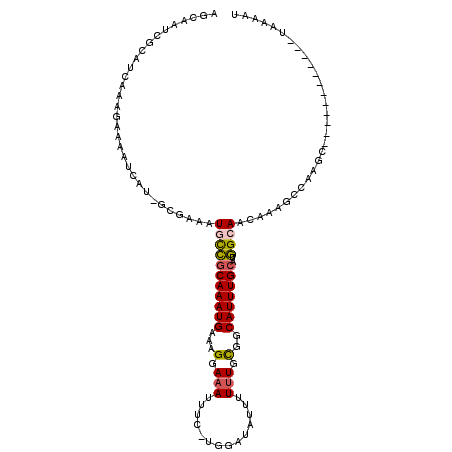
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.32 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
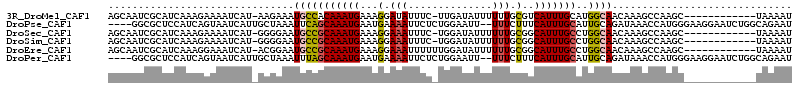
>3R_DroMel_CAF1 23206173 100 + 27905053 AGCAAUCGCAUCAAAGAAAAUCAU-AAGAAAUGCCACAAAUGAAAGGAUAUUUC-UUGAUAUUUUUUGCGUCAUUUGCAUGGCAACAAAGCCAAGC------------UAAAAU .((((.((((...(((((.((((.-.(((((((((..........)).))))))-))))).)))))))))....)))).((((......))))...------------...... ( -24.10) >DroPse_CAF1 3767 108 + 1 ----GGCGCUCCAUCAGUAAUCAUUGCUAAAUUCAGCAAAUGAAUGAAAAUUCUCUGGAAUU--UUUCUUUCAUUUGCAUUGCAGAUAAACCAUGGGAAGGAAUCUGGCAGAAU ----...................((((((.((((.(((((((((.((((((((....)))))--)))..))))))))).((.((.........)).))..)))).))))))... ( -26.80) >DroSec_CAF1 3839 100 + 1 AGCAAUCGCAUCAAAGAAAAUCAU-GGGGAAUGCCGCAAAUGAAAGGAAAUUUC-UGGAUAUUUUUUGCGGCAUUUGCCUGGCAACAAAGCCAAGC------------UAAAAU (((.....................-((.((((((((((((.....(((....))-)........)))))))))))).))((((......)))).))------------)..... ( -30.92) >DroSim_CAF1 3851 100 + 1 AGCAAUCGCAUCAAAGAAAAUCAU-GGGGAAUGCCGCAAAUGAAAGGAAAUUUC-UGGAUAUUUUUUGCGGCAUUUGCCUGGCAACAAAGCCAAGC------------UAAAAU (((.....................-((.((((((((((((.....(((....))-)........)))))))))))).))((((......)))).))------------)..... ( -30.92) >DroEre_CAF1 3715 101 + 1 AGCAAUCGCAUCAAAGGAAAUCAU-ACGGAAUGCCGCAAAUGAAAGGAAAUUUUUUGGAUAUUUUUUGCGGCAUUUGCCUGGCAACAAAGCCAAGC------------UAAAAU .((....))...............-...((((((((((((........((....))........))))))))))))((.((((......)))).))------------...... ( -25.79) >DroPer_CAF1 3779 108 + 1 ----GGCGCUCCAUCAGUAAUCAUUGCUAAAUUUAGCAAAUGAAUGAAAAUUCUCUGGAAUU--UUUCUUUCAUUUGCAUUGCAGAUAAACCAUGGGAAGGAAUCUGGCAGAAU ----...................((((........(((((((((.((((((((....)))))--)))..)))))))))....(((((...((.......)).)))))))))... ( -25.50) >consensus AGCAAUCGCAUCAAAGAAAAUCAU_GCGAAAUGCCGCAAAUGAAAGGAAAUUUC_UGGAUAUUUUUUGCGGCAUUUGCAUGGCAACAAAGCCAAGC____________UAAAAU ...............................(((((((((((...(.(((..............))).)..)))))))..)))).............................. ( -8.46 = -8.32 + -0.14)
| Location | 23,206,173 – 23,206,273 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.98 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.53 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
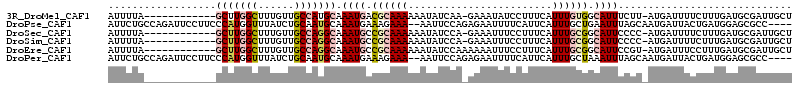
>3R_DroMel_CAF1 23206173 100 - 27905053 AUUUUA------------GCUUGGCUUUGUUGCCAUGCAAAUGACGCAAAAAAUAUCAA-GAAAUAUCCUUUCAUUUGUGGCAUUUCUU-AUGAUUUUCUUUGAUGCGAUUGCU ......------------((.((((......)))).))......((((..(((.(((((-(((((..((........).)..)))))).-.))))....)))..))))...... ( -20.90) >DroPse_CAF1 3767 108 - 1 AUUCUGCCAGAUUCCUUCCCAUGGUUUAUCUGCAAUGCAAAUGAAAGAAA--AAUUCCAGAGAAUUUUCAUUCAUUUGCUGAAUUUAGCAAUGAUUACUGAUGGAGCGCC---- ....((((((((.((.......))...)))))....(((((((((.((((--(.(((....)))))))).)))))))))........)))....................---- ( -23.10) >DroSec_CAF1 3839 100 - 1 AUUUUA------------GCUUGGCUUUGUUGCCAGGCAAAUGCCGCAAAAAAUAUCCA-GAAAUUUCCUUUCAUUUGCGGCAUUCCCC-AUGAUUUUCUUUGAUGCGAUUGCU ......------------(((((((......))))))).(((((((((((.........-..............)))))))))))....-...............((....)). ( -26.80) >DroSim_CAF1 3851 100 - 1 AUUUUA------------GCUUGGCUUUGUUGCCAGGCAAAUGCCGCAAAAAAUAUCCA-GAAAUUUCCUUUCAUUUGCGGCAUUCCCC-AUGAUUUUCUUUGAUGCGAUUGCU ......------------(((((((......))))))).(((((((((((.........-..............)))))))))))....-...............((....)). ( -26.80) >DroEre_CAF1 3715 101 - 1 AUUUUA------------GCUUGGCUUUGUUGCCAGGCAAAUGCCGCAAAAAAUAUCCAAAAAAUUUCCUUUCAUUUGCGGCAUUCCGU-AUGAUUUCCUUUGAUGCGAUUGCU ......------------(((((((......))))))).(((((((((((........................))))))))))).(((-((.(.......).)))))...... ( -30.66) >DroPer_CAF1 3779 108 - 1 AUUCUGCCAGAUUCCUUCCCAUGGUUUAUCUGCAAUGCAAAUGAAAGAAA--AAUUCCAGAGAAUUUUCAUUCAUUUGCUAAAUUUAGCAAUGAUUACUGAUGGAGCGCC---- ....((((((((.((.......))...)))))....(((((((((.((((--(.(((....)))))))).)))))))))........)))....................---- ( -23.10) >consensus AUUUUA____________GCUUGGCUUUGUUGCCAGGCAAAUGACGCAAAAAAUAUCCA_AAAAUUUCCUUUCAUUUGCGGCAUUCCGC_AUGAUUUCCUUUGAUGCGAUUGCU ..................(((((((......))))))).((((.((((((........................)))))).))))............................. ( -9.77 = -10.53 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:22 2006