
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,203,151 – 23,203,247 |
| Length | 96 |
| Max. P | 0.891864 |

| Location | 23,203,151 – 23,203,247 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.87 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
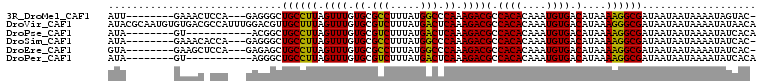
>3R_DroMel_CAF1 23203151 96 + 27905053 -GUACUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUC---UGGAGUUUC--------AAU -...................(((((...(((((((....))))(((((((((.....))))))))))))....)))))(((((..---.)).)))..--------... ( -22.50) >DroVir_CAF1 763 108 + 1 UGUUAUAUUUUAUUAUUAUCGCCCUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAAGCAACGUCCAAAUGGCGUCACACAUUGCGUAU ..........................((((.((....(((((((((((((((.(((.....)))((..........))......)))).))))))))))).)))))). ( -25.00) >DroPse_CAF1 754 89 + 1 UGUGAUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAGGCAGCCGU-----------AC--------UAU .((((((......)))))).(((((...(((((((....))))(((((((((.....))))))))))))....)))))......-----------..--------... ( -23.50) >DroSim_CAF1 839 96 + 1 -GUGAUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUC---UGGUGUUUC--------UAU -((((((......)))))).(((((...(((((((....))))(((((((((.....))))))))))))....))))).(((...---.))).....--------... ( -25.30) >DroEre_CAF1 813 96 + 1 -GUGAUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCUCUC---UGGAGCUUC--------UAC -((((((......)))))).(((((...(((((((....))))(((((((((.....))))))))))))....)))))(((((..---..)))))..--------... ( -29.90) >DroPer_CAF1 778 89 + 1 UGUGAUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAGGCAGCCCU-----------AC--------UAU .((((((......)))))).(((((...(((((((....))))(((((((((.....))))))))))))....)))))......-----------..--------... ( -23.50) >consensus _GUGAUAUUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGCCAUAAAGACGCACAAACUAAGGCAGCCCUC___UGG_GUUAC________UAU .((((((......)))))).(((((...(((((((....))))(((((((((.....))))))))))))....))))).............................. (-21.56 = -21.87 + 0.31)



| Location | 23,203,151 – 23,203,247 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -18.38 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23203151 96 - 27905053 AUU--------GAAACUCCA---GAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAGUAC- ...--------.(((((...---((((....)))))))))...(((((((.((((........))))(((....))).....)))))))..................- ( -21.70) >DroVir_CAF1 763 108 - 1 AUACGCAAUGUGUGACGCCAUUUGGACGUUGCUUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAGGGCGAUAAUAAUAAAAUAUAACA .(((((...))))).((((.((((...((..(.((.((.((.((((((((.......)))))))))).)).)).)..)).)))).))))................... ( -27.70) >DroPse_CAF1 754 89 - 1 AUA--------GU-----------ACGGCUGCCUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAUCACA ...--------..-----------......(((((....(((((((((((.......))))))))((((....)))))))...)))))((((.........))))... ( -20.80) >DroSim_CAF1 839 96 - 1 AUA--------GAAACACCA---GAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAUCAC- ...--------((.......---((((....))))...((((.(((((((.((((........))))(((....))).....)))))))...)))).......))..- ( -21.10) >DroEre_CAF1 813 96 - 1 GUA--------GAAGCUCCA---GAGAGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAUCAC- ...--------..(((((..---..)))))(((((.((((.((.(((.....))).)).))))(.((((....)))).)....)))))((((.........))))..- ( -25.40) >DroPer_CAF1 778 89 - 1 AUA--------GU-----------AGGGCUGCCUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAUCACA ...--------..-----------......(((((....(((((((((((.......))))))))((((....)))))))...)))))((((.........))))... ( -20.80) >consensus AUA________GAAAC_CCA___GAGGGCUGCCUUAGUUUGUGCGCCUUUAUGACCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAAUAUCAC_ .............................((((((.((((.((.(((.....))).)).))))(.((((....)))).)....))))))................... (-18.38 = -17.60 + -0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:19 2006