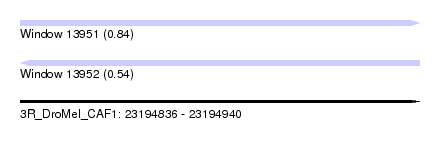
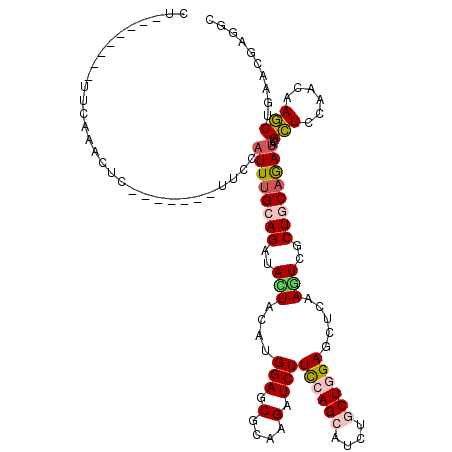
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,194,836 – 23,194,940 |
| Length | 104 |
| Max. P | 0.835670 |

| Location | 23,194,836 – 23,194,940 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23194836 104 + 27905053 CU--------UUCAAUCUC-------CUUCAUUCGCAGAUAUUACAUGGAGCGCAAGAUCUUCCAGCAUCUCCUGGAGCUCAAGUCGCUGCAGAUUCGCUCCAGUAAGCUGAACGAGGC ..--------........(-------(((......(((...((((.(((((((...((((((((((......)))))((..........)))))))))))))))))).)))...)))). ( -30.10) >DroVir_CAF1 10353 107 + 1 UU--------UGCAUGCUCUU----AUACUCUUUGCAGAUACUACAUGGAGCGCAAGAUCUUCCAGCAUCUGCUGGAGCUCAAGUCGCUACAAAUACGCUCAAACAAGCUGAACGAAGC ((--------((((.......----........)))))).........(((((........((((((....))))))((.......))........)))))......(((......))) ( -24.06) >DroGri_CAF1 11050 100 + 1 CU--------GCCAU-----------CCCUAUUUGCAGAUACUACAUGGAGCGCAAGAUCUUUCAGCAUCUGCUUGAGCUCAAGUCGCUGCAGAUACGCUCCAACAAGCUGAACGAGGC ..--------.....-----------.(((.....(((........(((((((....((((..((((....(((((....))))).)))).)))).))))))).....)))....))). ( -28.12) >DroWil_CAF1 38448 113 + 1 CC------UCUUGAAAUUUAUGUUUGUUUCAUUUGCAGAUAUUACAUGGAGCGCAAGAUCUUUCAGCAUUUGCUGGAGCUGAAAUCCCUACAGAUACGUUCCAGUAAACUGAAUGAGUC ..------(((((...(((((((.((((((....).)))))..)))))))...)))))((.(((((..(((((((((((....(((......)))..)))))))))))))))).))... ( -31.60) >DroMoj_CAF1 10849 107 + 1 CU--------GGCACACUUCA----CUUCUAUUUGCAGAUACUACAUGGAGCGCAAGAUCUUCCAGCAUCUGCUGGAGCUCAAGUCGCUGCAAAUACGCUCCAACAAGCUGAACGAGGU ..--------.....(((((.----....(((((((((..(((....(((.(....).)))((((((....)))))).....)))..))))))))).(((......))).....))))) ( -31.90) >DroAna_CAF1 8546 114 + 1 CCUACUUUG-UUUAAUUUCUC----GUUUCAUUUGCAGAUACUACAUGGAACGAAAGAUCUUCCAGCAUCUUCUCGAGCUUAAAUCUCUGCAGAUCCGCUCCAGCAAGCUCAACGAGGC .........-.......((((----(((..(((((((((.......((((((....)...)))))((.((.....)))).......)))))))))..(((......)))..))))))). ( -25.90) >consensus CU________UUCAAACUC_______UUCCAUUUGCAGAUACUACAUGGAGCGCAAGAUCUUCCAGCAUCUGCUGGAGCUCAAGUCGCUGCAGAUACGCUCCAACAAGCUGAACGAGGC ..............................((((((((..(((....(((.(....).)))((((((....)))))).....)))..))))))))..(((......))).......... (-17.00 = -17.50 + 0.50)

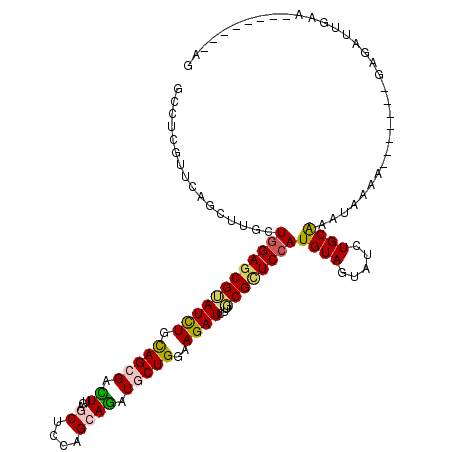
| Location | 23,194,836 – 23,194,940 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23194836 104 - 27905053 GCCUCGUUCAGCUUACUGGAGCGAAUCUGCAGCGACUUGAGCUCCAGGAGAUGCUGGAAGAUCUUGCGCUCCAUGUAAUAUCUGCGAAUGAAG-------GAGAUUGAA--------AG .((((((((.(((((((((((((.((((.(((((.((((.....))))...)))))..))))....))))))).)))).....))))))).))-------)........--------.. ( -34.70) >DroVir_CAF1 10353 107 - 1 GCUUCGUUCAGCUUGUUUGAGCGUAUUUGUAGCGACUUGAGCUCCAGCAGAUGCUGGAAGAUCUUGCGCUCCAUGUAGUAUCUGCAAAGAGUAU----AAGAGCAUGCA--------AA ....(((((((.....)))))))..((((((((.........((((((....))))))...(((((..(((..(((((...)))))..)))..)----)))))).))))--------)) ( -34.90) >DroGri_CAF1 11050 100 - 1 GCCUCGUUCAGCUUGUUGGAGCGUAUCUGCAGCGACUUGAGCUCAAGCAGAUGCUGAAAGAUCUUGCGCUCCAUGUAGUAUCUGCAAAUAGGG-----------AUGGC--------AG (((((((((((.((((((.((.....)).)))))).))))))....((((((((((...((........))....)))))))))).......)-----------).)))--------.. ( -34.50) >DroWil_CAF1 38448 113 - 1 GACUCAUUCAGUUUACUGGAACGUAUCUGUAGGGAUUUCAGCUCCAGCAAAUGCUGAAAGAUCUUGCGCUCCAUGUAAUAUCUGCAAAUGAAACAAACAUAAAUUUCAAGA------GG ..(((...(((.((((((((.((((.....(((..(((((((..........)))))))..))))))).)))).))))...)))....(((((..........))))).))------). ( -27.70) >DroMoj_CAF1 10849 107 - 1 ACCUCGUUCAGCUUGUUGGAGCGUAUUUGCAGCGACUUGAGCUCCAGCAGAUGCUGGAAGAUCUUGCGCUCCAUGUAGUAUCUGCAAAUAGAAG----UGAAGUGUGCC--------AG ..((.((...((((..(((((((((....(.((.......))((((((....)))))).)....)))))))))(((((...)))))........----..))))..)).--------)) ( -32.60) >DroAna_CAF1 8546 114 - 1 GCCUCGUUGAGCUUGCUGGAGCGGAUCUGCAGAGAUUUAAGCUCGAGAAGAUGCUGGAAGAUCUUUCGUUCCAUGUAGUAUCUGCAAAUGAAAC----GAGAAAUUAAA-CAAAGUAGG ..((((((..(.((((..((((.(((((....)))))...))))....(((((((((((((....)).))))....))))))))))).)..)))----)))........-......... ( -31.70) >consensus GCCUCGUUCAGCUUGCUGGAGCGUAUCUGCAGCGACUUGAGCUCCAGCAGAUGCUGGAAGAUCUUGCGCUCCAUGUAGUAUCUGCAAAUAAAA_______GAGAUUGAA________AG ................((((((((((((.(((((.((...((....)))).)))))..))))...))))))))((((.....))))................................. (-17.49 = -17.38 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:16 2006