
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,188,949 – 23,189,144 |
| Length | 195 |
| Max. P | 0.998381 |

| Location | 23,188,949 – 23,189,053 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23188949 104 + 27905053 AGGAGGCCGCAAAGCCA---CCAAGUGCCGCAGAACAACCACAACCUAAUGCCAACACAGUGGAAGGGAUGGAAUCUGCGGCGGCAGAGCCAGCGUCUGUUGGCAAA .((.(((......))).---))..((((((((((....(((...(((....(((......))).)))..)))..)))))))).))...((((((....))))))... ( -40.10) >DroSec_CAF1 3893 100 + 1 ----GGCCGCAAAGCCA---CCAAGUGCCGCAGAACAACCACAGCCUAAUGCCAACACGGUGGAAGGGAUGGAAUCUGCGGCUGCAGAGCCAGCGUCUGUUGGCAAA ----(((......))).---....((((((((((....(((...(((..((((.....))))..)))..)))..)))))))).))...((((((....))))))... ( -38.40) >DroSim_CAF1 4158 104 + 1 AGGAGGCCACAAAGCCA---CCAAGUGCCGCAGAACAACCACCGCCUAAUGCCAACACGGUGGAAGGGAUGGAAUCUGCGGCUGCAGAGCCAGCGUCUGUUGGCAAA .((.(((......))).---))..((((((((((.((.((.(((((............)))))...)).))...)))))))).))...((((((....))))))... ( -43.10) >DroEre_CAF1 4146 104 + 1 AGGAGGCCGCAAAGCCA---CCAAGUGCCGCAGAGCAACCACAGCCUAAUGCCAACACGGUCGAAGGGGCGGAAUCUGUGGCUACAGAGCCAGCUUCUGUUGGCAAA .((.(((......))).---))..((((((((((....((...((((...(((.....))).....))))))..)))))))).))...((((((....))))))... ( -41.30) >DroYak_CAF1 3500 107 + 1 AGGAAGCCGCAAAGCCACCACCAAGUGCCGCAGAACAACCACAGCCUAAUGCCAACAAGGCCGACGGGAUGGAAUCUGCGGCUGUGGAGCCAGUGUCUGUUGGCAAA .((..((......))..)).(((...((((((((....(((...(((...(((.....)))....))).)))..))))))))..))).(((((......)))))... ( -38.10) >consensus AGGAGGCCGCAAAGCCA___CCAAGUGCCGCAGAACAACCACAGCCUAAUGCCAACACGGUGGAAGGGAUGGAAUCUGCGGCUGCAGAGCCAGCGUCUGUUGGCAAA ....(((......)))........((((((((((....(((...(((..((((.....))))...))).)))..)))))))).))...((((((....))))))... (-33.40 = -33.60 + 0.20)



| Location | 23,188,949 – 23,189,053 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -39.26 |
| Consensus MFE | -31.42 |
| Energy contribution | -31.18 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23188949 104 - 27905053 UUUGCCAACAGACGCUGGCUCUGCCGCCGCAGAUUCCAUCCCUUCCACUGUGUUGGCAUUAGGUUGUGGUUGUUCUGCGGCACUUGG---UGGCUUUGCGGCCUCCU ...((((........))))......((((((((..((((.(((.(((......)))....)))..))))....))))))))....((---.((((....)))).)). ( -37.90) >DroSec_CAF1 3893 100 - 1 UUUGCCAACAGACGCUGGCUCUGCAGCCGCAGAUUCCAUCCCUUCCACCGUGUUGGCAUUAGGCUGUGGUUGUUCUGCGGCACUUGG---UGGCUUUGCGGCC---- ...(((((((((.(....)))))..((((((((..((((.(((.(((......)))....)))..))))....))))))))..))))---)((((....))))---- ( -35.40) >DroSim_CAF1 4158 104 - 1 UUUGCCAACAGACGCUGGCUCUGCAGCCGCAGAUUCCAUCCCUUCCACCGUGUUGGCAUUAGGCGGUGGUUGUUCUGCGGCACUUGG---UGGCUUUGUGGCCUCCU ...((((........))))......((((((((..((((((((.(((......)))....))).)))))....))))))))....((---.((((....)))).)). ( -42.50) >DroEre_CAF1 4146 104 - 1 UUUGCCAACAGAAGCUGGCUCUGUAGCCACAGAUUCCGCCCCUUCGACCGUGUUGGCAUUAGGCUGUGGUUGCUCUGCGGCACUUGG---UGGCUUUGCGGCCUCCU ..((((((((((((..((((((((....)))))....))).)))).....))))))))....((((..(.....)..))))....((---.((((....)))).)). ( -38.20) >DroYak_CAF1 3500 107 - 1 UUUGCCAACAGACACUGGCUCCACAGCCGCAGAUUCCAUCCCGUCGGCCUUGUUGGCAUUAGGCUGUGGUUGUUCUGCGGCACUUGGUGGUGGCUUUGCGGCUUCCU ...((((........)))).....(((((((((..((((((((...(((.....))).....((((..(.....)..))))...))).))))).))))))))).... ( -42.30) >consensus UUUGCCAACAGACGCUGGCUCUGCAGCCGCAGAUUCCAUCCCUUCCACCGUGUUGGCAUUAGGCUGUGGUUGUUCUGCGGCACUUGG___UGGCUUUGCGGCCUCCU ...((((........))))......((((((((..((((.(((....((.....))....)))..))))....))))))))..........((((....)))).... (-31.42 = -31.18 + -0.24)


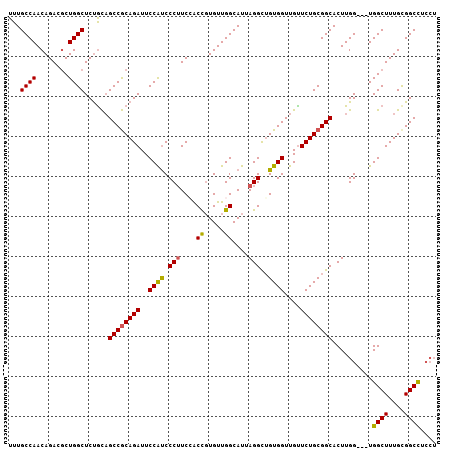
| Location | 23,189,053 – 23,189,144 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.60 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.91 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23189053 91 - 27905053 GACCGGGUGGAG------------CAGCGGUGGGCGGAGGUAUGGGAGUAGGAUUGGUAUUUGGUUUCGGCUUGGCUUCAGCCAGCUCCAUCUGUUCUGUUUU ((.(((((((((------------(...(((.(((.(((...((..(.((((.......)))).)..)).))).)))...))).)))))))))).))...... ( -31.20) >DroSec_CAF1 3993 91 - 1 GAGCGGGUGGAA------------CAGCGGUGGGCGGAGGUGUGGGAGUAGGAUUGGUAUUUGGUUUAGGCUUGGCUUCAGCCUGCUCCAUCUGUUCGGUUUU (((((((((((.------------.....(..(((.(((((....((((.((((..(...)..))))..)))).))))).)))..))))))))))))...... ( -32.80) >DroSim_CAF1 4262 91 - 1 GAGCGGGUGGAG------------CAGCGGUGGGCGGAGGUGUGGGAGUAGGAUUGGUAUUUGGUUUCGGCUUGGCUUCAGCCUGCUCCAUCUGUUCGGUUUU ((((((((((((------------(((.(.(((((.((...((.((((..((((....))))..)))).)))).)).))).))))))))))))))))...... ( -39.40) >DroEre_CAF1 4250 103 - 1 GAGCAGGUGGAGCAGCGGCUGGAGCAGCGGUGGGUGGAGGUGUGGGACUAGGACUGGUAUUUGGUUUCGGCAUGACUGCAGCCUGCUCCAUCUGCUCGGUUUU ((((((((((((((((.((((...)))).))((.((.(((((((..((((((.......))))))..).))))..)).)).))))))))))))))))...... ( -49.00) >consensus GAGCGGGUGGAG____________CAGCGGUGGGCGGAGGUGUGGGAGUAGGAUUGGUAUUUGGUUUCGGCUUGGCUUCAGCCUGCUCCAUCUGUUCGGUUUU (((((((((((..................((((((.(((((....((((.((((..(...)..))))..)))).))))).)))))))))))))))))...... (-28.53 = -28.91 + 0.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:13 2006