
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,185,190 – 23,185,288 |
| Length | 98 |
| Max. P | 0.652721 |

| Location | 23,185,190 – 23,185,288 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.14 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -8.74 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23185190 98 + 27905053 --------CAAGUCC-UUGGCAACUCACCACGUUGCCCAGGACAACGGCUUCCAAUUCUGCCC---GCAG-GACCCUGCACC---CUCUUACCCUCUCGUUGGCUCCUUCCAGG --------...((((-(.((((((.......)))))).)))))...(((..........))).---((((-....))))...---..............((((......)))). ( -30.00) >DroPse_CAF1 330 107 + 1 AGUUUCUCCAAGUGCAGUGGUAACUCACCACGUUGCCCAUAACAACCGUUUUCCAACCCGCUUUUGGCCG-UUUCCAGGCGU--CGCGUCCGACACUU----GCACUUUCCAGG .........((((((((((((.....)))))((((.....(((....)))...)))).........(((.-......)))((--((....))))...)----))))))...... ( -25.00) >DroSec_CAF1 128 102 + 1 --------CAAGUCC-UUGGCAACUCACCACGUUGCCCAGGACAACGGCUUCCAAUCCUGCCC---GCAGGGACCCUGCCCCCCCCUCUCCACCAAUCGAUGGCUACUUCCCGG --------...((((-(.((((((.......)))))).)))))...(((..........))).---...((((....(((.....................)))....)))).. ( -30.30) >DroSim_CAF1 404 100 + 1 --------CAAGUCC-UUGGCAACUCACCACGUUGCCCAGAACAACGGCUUCCAAUCCUGCCC---GCAG-GACCCUGCCCC-CCCUCUCCACCACUCGAUGGCUACUUCCAGG --------..(((((-(.((((((.......)))))).))......(((......(((((...---.)))-))....)))..-..................))))......... ( -23.60) >DroEre_CAF1 395 84 + 1 -----CU-CCAGUCC-UUGGCAACUCACCACGUUGCCCAGGACAACGACUUCCAAUCCUUCUC---GCAG-GACACACCCAC-C-------------CG-----CCCUUCCAGG -----..-((.((((-(.((((((.......)))))).)))))............((((....---..))-)).........-.-------------..-----........)) ( -21.70) >DroPer_CAF1 476 107 + 1 AGUUUCUCCAAGUGCAGUGGUAACUCACCACGUUGCCCAUAACAACCGUUUUCCAACCCGCUUUUGGCCG-UUUCCAGGCGU--CGCGUCCGACACUU----GCACUUUCCAGG .........((((((((((((.....)))))((((.....(((....)))...)))).........(((.-......)))((--((....))))...)----))))))...... ( -25.00) >consensus ________CAAGUCC_UUGGCAACUCACCACGUUGCCCAGAACAACGGCUUCCAAUCCUGCCC___GCAG_GACCCAGCCCC__CCUCUCCACCACUCG___GCUCCUUCCAGG ...........(((....((((((.......))))))....................((((.....)))).)))........................................ ( -8.74 = -8.68 + -0.05)

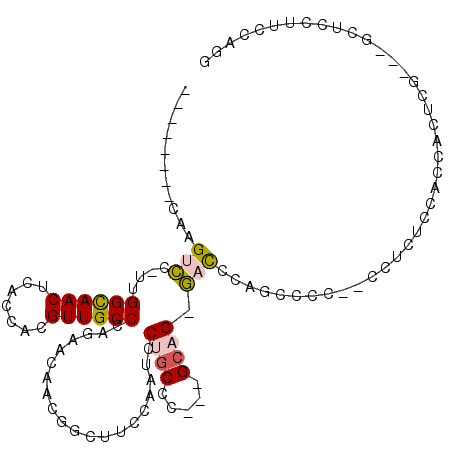
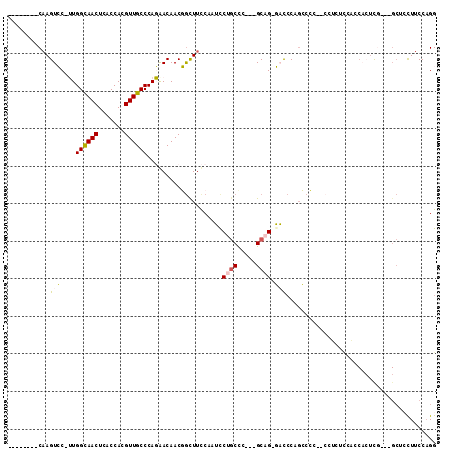
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:07 2006