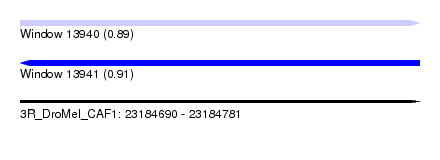
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,184,690 – 23,184,781 |
| Length | 91 |
| Max. P | 0.914901 |

| Location | 23,184,690 – 23,184,781 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
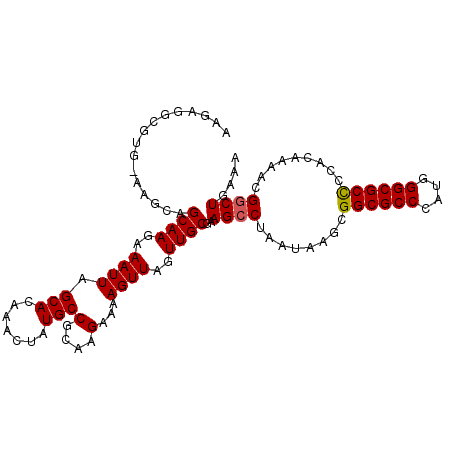
>3R_DroMel_CAF1 23184690 91 + 27905053 ---------------AGCAAGAAAUUAGCACAAACUAUGCCGCAAGAAAAGUUAGUUGCGAAGCCUAAUAAGCGGCGCCCAUGGGCGCUCCACAAAACGGCUGAAA ---------------.((((..((((.(((.......)))(....)...))))..))))..((((........((((((....)))))).........)))).... ( -23.63) >DroSec_CAF1 27561 106 + 1 AAGAGGCGUGGAAGCAGCAAGAAAUUAGCACAAACUAUGCCGCAAGAAAAGUUAUUUGCGAAGCCUAAUAAGCGGCGCCCAUGGGCGCCCCACAAAACGGCUGGAA ....(((((((.....((((....)).)).....)))))))(((((........)))))..((((........((((((....)))))).........)))).... ( -34.13) >DroEre_CAF1 32103 106 + 1 AAGAGGCGUGCAAGCGGCAAGAAAUUAGCACAAACUAUGCCGCAAGAAAAGUUAGUUGCGAAGCCUAAUAAGCGGCGCCCAUGGGCGCCCCACAAAGCGACUGCAC .......((((..((((((......(((......)))))))))..........((((((...((.......))((((((....)))))).......)))))))))) ( -38.00) >consensus AAGAGGCGUG_AAGCAGCAAGAAAUUAGCACAAACUAUGCCGCAAGAAAAGUUAGUUGCGAAGCCUAAUAAGCGGCGCCCAUGGGCGCCCCACAAAACGGCUGAAA ................((((..((((.(((.......)))(....)...))))..))))..((((........((((((....)))))).........)))).... (-22.42 = -22.53 + 0.11)

| Location | 23,184,690 – 23,184,781 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -27.73 |
| Energy contribution | -27.62 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
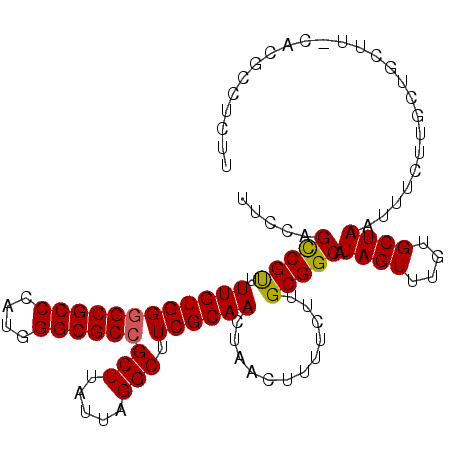
>3R_DroMel_CAF1 23184690 91 - 27905053 UUUCAGCCGUUUUGUGGAGCGCCCAUGGGCGCCGCUUAUUAGGCUUCGCAACUAACUUUUCUUGCGGCAUAGUUUGUGCUAAUUUCUUGCU--------------- .....(((((.((((((((((((....))))).(((.....))))))))))............))))).((((....))))..........--------------- ( -27.02) >DroSec_CAF1 27561 106 - 1 UUCCAGCCGUUUUGUGGGGCGCCCAUGGGCGCCGCUUAUUAGGCUUCGCAAAUAACUUUUCUUGCGGCAUAGUUUGUGCUAAUUUCUUGCUGCUUCCACGCCUCUU ....((.((((((((((((((((....))))))(((.....))).)))))))...........((((((((((....))))......))))))....))).))... ( -33.10) >DroEre_CAF1 32103 106 - 1 GUGCAGUCGCUUUGUGGGGCGCCCAUGGGCGCCGCUUAUUAGGCUUCGCAACUAACUUUUCUUGCGGCAUAGUUUGUGCUAAUUUCUUGCCGCUUGCACGCCUCUU ((((((.....((((((((((((....))))))(((.....))).))))))............((((((((((....))))......))))))))))))....... ( -40.50) >consensus UUCCAGCCGUUUUGUGGGGCGCCCAUGGGCGCCGCUUAUUAGGCUUCGCAACUAACUUUUCUUGCGGCAUAGUUUGUGCUAAUUUCUUGCUGCUU_CACGCCUCUU .....(((((.((((((((((((....))))))(((.....))).))))))............))))).((((....))))......................... (-27.73 = -27.62 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:06 2006