| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,180,992 – 23,181,113 |
| Length | 121 |
| Max. P | 0.584062 |

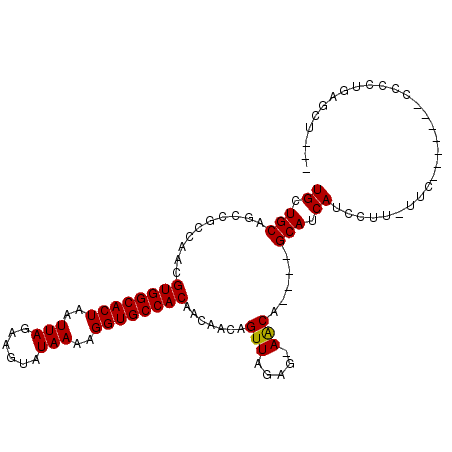
| Location | 23,180,992 – 23,181,087 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23180992 95 + 27905053 CUGCAGUGAACGCAGCCAA-GCGCCAUUGUUGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAGAAGC ((((((..(.(((......-)))......)..))))))........((((((((..(((......)))..)))))))).................. ( -28.20) >DroVir_CAF1 31917 88 + 1 CUGCAAUGCAUGCAAC-------CCAUUGGUGUUGCAGCUACCAGCGUGGCACUAAUUAGAAGUAUAAAAGGUGCUACAACAACAUUUAAAG-AGC (((....((.((((((-------((...)).))))))))...))).((((((((..(((......)))..))))))))..............-... ( -24.30) >DroPse_CAF1 19078 90 + 1 CGGCAAUGUAUGCAGCC-----GCCAUUGUUGCUGCAGCCGGCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AAC ((((......((((((.-----((....)).)))))))))).....((((((((..(((......)))..))))))))..............-... ( -29.70) >DroMoj_CAF1 39497 88 + 1 CUGCAAUGCAUGCAAG-------CCAUUGUUGUUGCAGCUACCAAUGUGGCACUAAUUAGAAGUAUAAAAGGUGCUACAACAACAGUUUAAG-AGC (((((((....((((.-------...)))).))))))).......(((((((((..(((......)))..))))))))).............-... ( -22.30) >DroAna_CAF1 26049 95 + 1 AUGCAAUGCAUGCUGCCGCCCCGCCAUUGUUGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AAC .......((..(((((.((..((....))..)).))))).))....((((((((..(((......)))..))))))))..............-... ( -24.80) >DroPer_CAF1 21778 90 + 1 CGGCAAUGUGUGCAGCC-----ACCAUUGUUGCUGCAGCCGGCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AAC ((((......((((((.-----((....)).)))))))))).....((((((((..(((......)))..))))))))..............-... ( -29.40) >consensus CUGCAAUGCAUGCAGCC_____GCCAUUGUUGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG_AAC ..((..((((.(((((............))))))))))).......((((((((..(((......)))..)))))))).................. (-19.31 = -19.12 + -0.19)

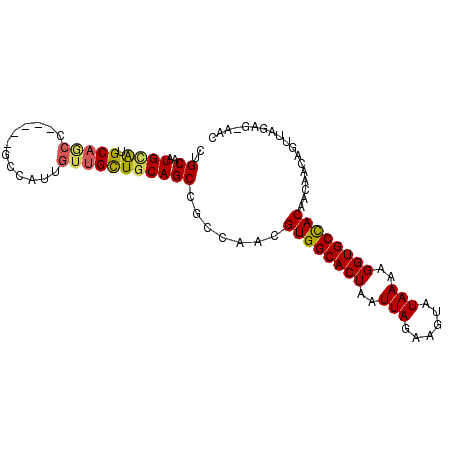

| Location | 23,180,992 – 23,181,087 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -21.29 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23180992 95 - 27905053 GCUUCUCUAACUGUUGUUGUGGCACCUUUUAUACUUCUAAUUAGUGCCACGUUGGCGGCUGCAGCAACAAUGGCGC-UUGGCUGCGUUCACUGCAG ..........(((((..((((((((...(((......)))...))))))))..)))))((((((......((((((-......))).))))))))) ( -29.10) >DroVir_CAF1 31917 88 - 1 GCU-CUUUAAAUGUUGUUGUAGCACCUUUUAUACUUCUAAUUAGUGCCACGCUGGUAGCUGCAACACCAAUGG-------GUUGCAUGCAUUGCAG (((-(......((.((((((((((((.....((((.......)))).......))).))))))))).))..))-------))((((.....)))). ( -22.60) >DroPse_CAF1 19078 90 - 1 GUU-CUCUAACUGUUGUUGUGGCACCUUUUAUACUUCUAAUUAGUGCCACGUUGCCGGCUGCAGCAACAAUGGC-----GGCUGCAUACAUUGCCG ...-......(((((((((((((((...(((......)))...)))))))(((((.....)))))))))))))(-----(((..........)))) ( -28.80) >DroMoj_CAF1 39497 88 - 1 GCU-CUUAAACUGUUGUUGUAGCACCUUUUAUACUUCUAAUUAGUGCCACAUUGGUAGCUGCAACAACAAUGG-------CUUGCAUGCAUUGCAG ...-.......(((((((((((((((.....((((.......)))).......))).))))))))))))....-------..((((.....)))). ( -25.80) >DroAna_CAF1 26049 95 - 1 GUU-CUCUAACUGUUGUUGUGGCACCUUUUAUACUUCUAAUUAGUGCCACGUUGGCGGCUGCAGCAACAAUGGCGGGGCGGCAGCAUGCAUUGCAU ((.-.(((..((((((((((.(((((......(((.......)))(((.....))))).))).))))))))))..)))..)).(((.....))).. ( -30.30) >DroPer_CAF1 21778 90 - 1 GUU-CUCUAACUGUUGUUGUGGCACCUUUUAUACUUCUAAUUAGUGCCACGUUGCCGGCUGCAGCAACAAUGGU-----GGCUGCACACAUUGCCG ...-.....((((((((((((((((...(((......)))...)))))))(((((.....))))))))))))))-----(((..........))). ( -29.70) >consensus GCU_CUCUAACUGUUGUUGUGGCACCUUUUAUACUUCUAAUUAGUGCCACGUUGGCGGCUGCAGCAACAAUGGC_____GGCUGCAUGCAUUGCAG ...........((((((((((((.........(((.......)))(((.....))).))))))))))))..............(((.....))).. (-21.29 = -20.98 + -0.30)

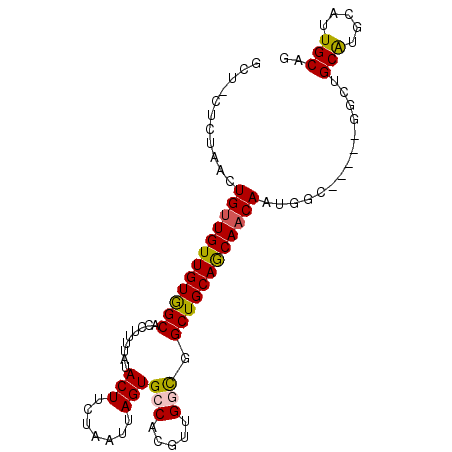

| Location | 23,181,021 – 23,181,113 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -18.03 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23181021 92 + 27905053 UGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAGAAGCA----GCAUCAUCCUUCUUC-------CCCCUGAGUG--- ((((((..........((((((((..(((......)))..))))))))(((....)))......)))----)))............-------..........--- ( -24.50) >DroPse_CAF1 19103 92 + 1 UGCUGCAGCCGGCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AACAGAGAGCAUCAUUCU---CC-------CAUCUGCUCU--- ....((((..((....((((((((..(((......)))..))))))))...........(((-((..((.....)).))))---))-------)..))))...--- ( -25.00) >DroEre_CAF1 27872 91 + 1 UGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AGCA----GCAUCAUCCUUGUUC-------CCCCUCAGUU--- ((((((..........((((((((..(((......)))..))))))))(((....)))....-.)))----)))............-------..........--- ( -24.50) >DroYak_CAF1 14564 98 + 1 UGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AGCAGC-AGCAUCAUCCUUGUUC-UUC--CCCCCUUACUU--- ((((((.(((......((((((((..(((......)))..))))))))(((....)))...)-.)).))-))))............-...--...........--- ( -26.30) >DroAna_CAF1 26079 101 + 1 UGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AACA----GCAUCAUCCUUCGUUCGUUCGCCCGCUUAGCCUGU .((((.(((.((.(((((((((((..(((......)))..)))))))).............(-(((.----............))))))).))..))))))).... ( -27.22) >DroPer_CAF1 21803 92 + 1 UGCUGCAGCCGGCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG-AACAGAGAGCAUCAUUCU---CU-------CAUCUGCUCU--- (((((....)))))..((((((((..(((......)))..))))))))..........((((-....(((((.......))---))-------).....))))--- ( -27.00) >consensus UGCUGCAGCCGCCAACGUGGCACUAAUUAGAAGUAUAAAAGGUGCCACAACAACAGUUAGAG_AACA____GCAUCAUCCUU_UUC_______CCCCUGAGCU___ ((.(((..........((((((((..(((......)))..)))))))).......(((.....))).....))).))............................. (-18.03 = -17.78 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:01 2006