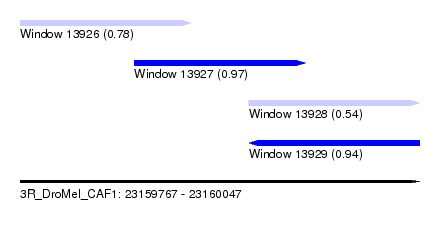
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,159,767 – 23,160,047 |
| Length | 280 |
| Max. P | 0.968403 |

| Location | 23,159,767 – 23,159,887 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.03 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23159767 120 + 27905053 AAGAAAAACCAGGCAAAAAAGUGAACAAUUUAAACUUUUGUAUGAAUAUAAAAGGGGGGCUGUGUGGGAAAUUAAAGCGCUGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGGCGAAAG ........((((.(....................((((((((....))))))))((((((.((((...........)))).)))..((((........)))).)))).))))........ ( -28.70) >DroSec_CAF1 9830 120 + 1 ACGAAAAACCAGGCAAAAAAGUGAACAAUUUAAACUUUUAUAUGUAUAUAAAAGGGGGGCUACGUAGGAAAUUAAAGCGCUGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGUCGAAAG .(((....((((.(....................((((((((....))))))))((((((..(((...........)))..)))..((((........)))).)))).)))).))).... ( -29.80) >DroSim_CAF1 15136 120 + 1 AAGAAAAACCAGGCAAAAAAGUGAACAAUUUAAACUUUUGUAUGAAUAAAUAAGGGGGGCUAUGCAGGAAAUUAAAGCGCUGCCAAGAUGUUUAAUAUCAUCACCUGGUUGGGUCGAAAG ..((..(((((((........((((((.......(((((((........))))))).(((..(((...........)))..)))....)))))).........)))))))...))..... ( -27.43) >consensus AAGAAAAACCAGGCAAAAAAGUGAACAAUUUAAACUUUUGUAUGAAUAUAAAAGGGGGGCUAUGUAGGAAAUUAAAGCGCUGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGUCGAAAG ........((((.(....................((((((((....))))))))((((((..(((...........)))..)))..((((........)))).)))).))))........ (-24.14 = -24.03 + -0.10)

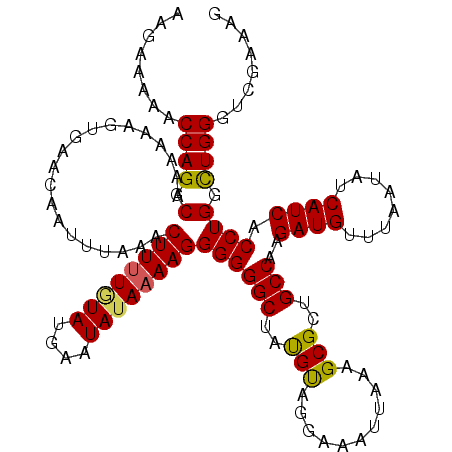

| Location | 23,159,847 – 23,159,967 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23159847 120 + 27905053 UGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGGCGAAAGAUCUAAAAUAAUUUUAUUUGCCACACAUUACCGCAGACAUGCUGGACUCCAUCCGCCAUUCUCGUCCUGCGAGUUGAACA ........(((((((..........((((.((((((((.((............)).))))))........(((((....))).)).......))))))..((((.....))))))))))) ( -26.30) >DroSec_CAF1 9910 120 + 1 UGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGUCGAAAGAUCUAAAAUAAUUUUAUUUGCCACACAUUACGGCAGACAUGCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUUGAACA ((((..((((........))))...((((((((((....))))))(((((....)))))))))........))))(((.((((((((................)))))))).)))..... ( -37.49) >DroSim_CAF1 15216 120 + 1 UGCCAAGAUGUUUAAUAUCAUCACCUGGUUGGGUCGAAAGAUCUAAAAUAAUUUUAUUUGCCACACAUUACGGCAGACAUGCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUCGAACA ((((..((((........))))...((((((((((....))))))(((((....)))))))))........))))(((.((((((((................)))))))).)))..... ( -37.79) >consensus UGCCAAGAUGUUUAAUAUCAUCACCUGGCUGGGUCGAAAGAUCUAAAAUAAUUUUAUUUGCCACACAUUACGGCAGACAUGCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUUGAACA ((((..((((........))))...((((((((((....))))))(((((....)))))))))........))))(((.((((((((................)))))))).)))..... (-32.40 = -32.96 + 0.56)

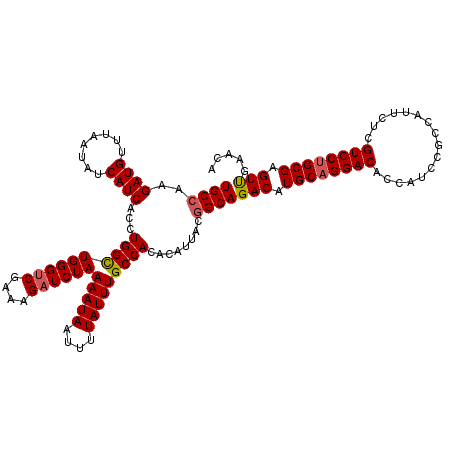
| Location | 23,159,927 – 23,160,047 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -30.36 |
| Energy contribution | -30.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23159927 120 + 27905053 GCUGGACUCCAUCCGCCAUUCUCGUCCUGCGAGUUGAACAAAACUUUUUACGCAUAGCCAGGACUCGCAGCGCAUUUUAGCUAAAAGCAUUUCAUUGCUGUUAACUAUUUUUGAUGGCGU ...(((.....)))((((((...((.(((((((((..........................))))))))).))....(((.(((.((((......)))).))).))).....)))))).. ( -30.87) >DroSec_CAF1 9990 120 + 1 GCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUUGAACAAAACUUUUUACGCAUAGCCAGGACUCGCAGCGCAUUUUAGCUAAAAGCAUUUCAACGCUGUUAACUAUUUUUGAUGGCGU ...(((.....)))((((((...((((((.(((((......))))).....((...))))))))..((((((.......((.....)).......))))))...........)))))).. ( -30.44) >DroSim_CAF1 15296 120 + 1 GCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUCGAACAAAACUUUUUACGCAUAGCCAGGACUCGCAGCGCAUUUUAGCUAAAAGCAUUUCAAUGCUGUUAACUAUUUUUGAUGGCGU ...(((.....)))((((((...((.(((((((((..........................))))))))).))....(((.(((.(((((....))))).))).))).....)))))).. ( -33.57) >consensus GCAGGACACCAUCCGCCAUUCUCGUCCUGCGAGUUGAACAAAACUUUUUACGCAUAGCCAGGACUCGCAGCGCAUUUUAGCUAAAAGCAUUUCAAUGCUGUUAACUAUUUUUGAUGGCGU ...(((.....)))((((((...((.(((((((((..........................))))))))).))....(((.(((.((((......)))).))).))).....)))))).. (-30.36 = -30.47 + 0.11)

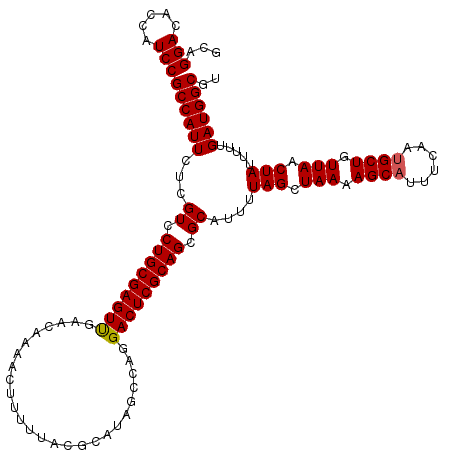
| Location | 23,159,927 – 23,160,047 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -36.71 |
| Energy contribution | -36.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23159927 120 - 27905053 ACGCCAUCAAAAAUAGUUAACAGCAAUGAAAUGCUUUUAGCUAAAAUGCGCUGCGAGUCCUGGCUAUGCGUAAAAAGUUUUGUUCAACUCGCAGGACGAGAAUGGCGGAUGGAGUCCAGC ..((((((.....(((((((.((((......)))).))))))).......((((((((...(((...((.......))...)))..)))))))).....).)))))((((...))))... ( -35.80) >DroSec_CAF1 9990 120 - 1 ACGCCAUCAAAAAUAGUUAACAGCGUUGAAAUGCUUUUAGCUAAAAUGCGCUGCGAGUCCUGGCUAUGCGUAAAAAGUUUUGUUCAACUCGCAGGACGAGAAUGGCGGAUGGUGUCCUGC ((((((((.....(((((((.(((((....))))).))))))).....((((.(..((((((((...))......((((......))))..))))))..)...))))))))))))..... ( -38.30) >DroSim_CAF1 15296 120 - 1 ACGCCAUCAAAAAUAGUUAACAGCAUUGAAAUGCUUUUAGCUAAAAUGCGCUGCGAGUCCUGGCUAUGCGUAAAAAGUUUUGUUCGACUCGCAGGACGAGAAUGGCGGAUGGUGUCCUGC ((((((((.....(((((((.(((((....))))).))))))).......(((((((((..(((...((.......))...))).))))))))).............))))))))..... ( -40.80) >consensus ACGCCAUCAAAAAUAGUUAACAGCAUUGAAAUGCUUUUAGCUAAAAUGCGCUGCGAGUCCUGGCUAUGCGUAAAAAGUUUUGUUCAACUCGCAGGACGAGAAUGGCGGAUGGUGUCCUGC ((((((((.....(((((((.(((((....))))).))))))).....((((.(..((((((((...))......((((......))))..))))))..)...))))))))))))..... (-36.71 = -36.93 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:55 2006