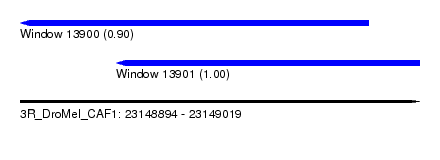
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,148,894 – 23,149,019 |
| Length | 125 |
| Max. P | 0.999606 |

| Location | 23,148,894 – 23,149,003 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.33 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23148894 109 - 27905053 CCCGCUUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUUGCGAAUGA-AAGUAGAGAGCGCCACUUACAAGUUAUGGUA-AACUCUUGGCCAGAAGUA .......((((((.(((((((((.(((((....)))))....((........)).....)-)))))))).(.((((.((((........)))-).....))))).)))))) ( -25.70) >DroSim_CAF1 4229 104 - 1 -----UUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUGA-AAGUAGAGAGCGCCACUCGCAACUUAUGGUU-AACUCUUGGCCAGGAGUA -----.........(((((((((.(((((....)))))....((((....)))).....)-)))))))).(((.....))).((((.(((((-(.....)))))).)))). ( -29.60) >DroYak_CAF1 4227 100 - 1 CCCG-----CUUUACUUUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUAAAAAGUAGAGAGCGCCA------GGAAAUGGUAAAACUCUUGGCCAAAAGUA ...(-----((((.(((((((((.(((((....)))))....((((....))))......))))))))).(.((((------(((...........)))))))).))))). ( -27.80) >consensus CCCG_UUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUGA_AAGUAGAGAGCGCCACU__CAAGUUAUGGUA_AACUCUUGGCCAGAAGUA .......((((((.((((((((..(((((....)))))....((........)).......)))))))).(.((((.......................))))).)))))) (-19.33 = -19.33 + 0.01)


| Location | 23,148,924 – 23,149,019 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23148924 95 - 27905053 AUUGGUGAAAAGGAAACCCGCUUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUUGCGAAUGA-AAGUAGAGAGCGCCACUUA ..(((((....((....))...........(((((((((.(((((....)))))....((........)).....)-))))))))..))))).... ( -24.60) >DroSim_CAF1 4259 74 - 1 ---------------------UUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUGA-AAGUAGAGAGCGCCACUCG ---------------------.........(((((((((.(((((....)))))....((((....)))).....)-))))))))........... ( -18.50) >DroYak_CAF1 4256 86 - 1 AUUGGCGGAAAGC-AACCCG-----CUUUACUUUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUAAAAAGUAGAGAGCGCCA---- ..(((((.(((((-.....)-----)))).(((((((((.(((((....)))))....((((....))))......)))))))))..)))))---- ( -27.40) >consensus AUUGG_G_AAAG__AACCCG_UUUACUUUACUCUACUUUAGUUGUCAAUGCAACAUAAGCUACUUAUGGCGAAUGA_AAGUAGAGAGCGCCACU__ ..............................((((((((..(((((....)))))....((........)).......))))))))........... (-14.72 = -14.50 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:28 2006