
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,144,345 – 23,144,455 |
| Length | 110 |
| Max. P | 0.645978 |

| Location | 23,144,345 – 23,144,455 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -38.61 |
| Consensus MFE | -24.38 |
| Energy contribution | -22.97 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
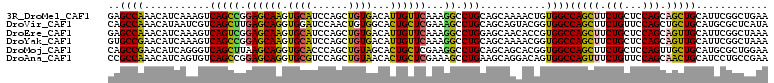
>3R_DroMel_CAF1 23144345 110 + 27905053 GAGCCAAACAUCAAAGUCAGCCGGAGCAAGUGCAUCCAGCUGUGACAUUGUUCAAAGGCCUGCAGCAAAACUGUGGCCAGCUUCUGCUCCAGCAGCUGCAUUCGGCUGAA ................(((((((((((....))...(((((((.............((((.((((.....))))))))(((....)))...)))))))..))))))))). ( -40.90) >DroVir_CAF1 1644 110 + 1 CAGCCAAACAUAAUCGUCAGCUUGAGCAGGUGGAUCCAACUGUGGCACUGCUCGAAAGCCUGCAGCAGUACGGUGGCCAGCUUCUGUUCCAGCUGCUGCAUGCGCUCAUA ...............((.(((((((((((.((...(((....)))))))))))))..((.(((((((((((((.((....)).))))....))))))))).))))).)). ( -38.10) >DroEre_CAF1 1378 110 + 1 GAGCCAAACAUCAAAGUCAGUCGGAGCAAGUGCAUCCAGCUGUGACAUUGUUCAAAGGCCUGGAGCAACACCGUGGCCAGCUUCUGCUCCAGCAGUUGCAUUCGGCUAAA .((((.((((.......((((.(((((....)).))).))))......))))....(((((((.......))).)))).((..((((....))))..))....))))... ( -36.62) >DroYak_CAF1 1388 110 + 1 GUGCCGAACAUCAAAGUCAGCCGGAGCAAGUGCAUCCAGCUGUGACAUUGUUCAAAGGCCUGCAGCAAAACGGUGGCCAGCUUCUGCUCCAGCAGUUGCAUUCGGCUAAA ..((((((((.......((((.(((((....)).))).))))......)))))...((((.((.(.....).)))))).((..((((....))))..))....))).... ( -38.22) >DroMoj_CAF1 1892 110 + 1 CAGCCGAACAUCAGGGUCAGCUUAAGCAGGUGCACCCAGCUGUAGCACUGCUCGAAGGCCUGCAGCAGCACGGUGGCCAGCUUCUGCUCCAGUUGCUGCAUGCGCUGGAA ((((.........(((((...(..(((((.(((((......)).))))))))..).)))))(((((((((.(.(((..(((....)))))).))))))).)))))))... ( -41.70) >DroAna_CAF1 1200 110 + 1 CCGCCAAACAUCAGUGUCAGCCGGAGCAGGUGCGUCCAGCUGUAACACUGCUCGAAAGCCUGAAGCAGGACAGUGGCCAGUUUCUGUUCCAGCAACUGCAUCCUGCCGAA .((.((.....((((....((.((((((((.((..(((.((((....(((((((......)).))))).)))))))...)).)))))))).)).)))).....)).)).. ( -36.10) >consensus CAGCCAAACAUCAAAGUCAGCCGGAGCAAGUGCAUCCAGCUGUGACACUGCUCAAAGGCCUGCAGCAAAACGGUGGCCAGCUUCUGCUCCAGCAGCUGCAUUCGGCUAAA ..((((...........(((((.((((((.((((......))))...))))))...)).)))...........))))(((((.(((...))).)))))............ (-24.38 = -22.97 + -1.41)


| Location | 23,144,345 – 23,144,455 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.19 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
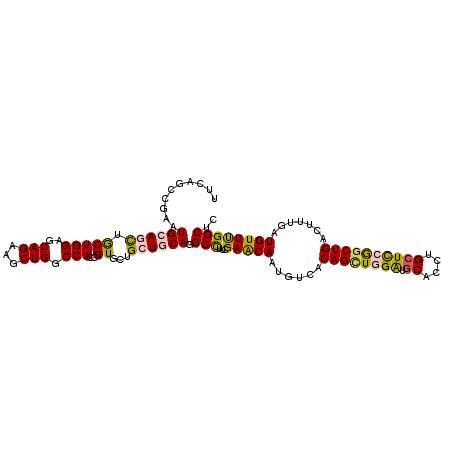
>3R_DroMel_CAF1 23144345 110 - 27905053 UUCAGCCGAAUGCAGCUGCUGGAGCAGAAGCUGGCCACAGUUUUGCUGCAGGCCUUUGAACAAUGUCACAGCUGGAUGCACUUGCUCCGGCUGACUUUGAUGUUUGGCUC ...((((((((((((((((....)))(((((((....))))))))))))...........(((.((((..((((((.((....)))))))))))).)))..)))))))). ( -45.00) >DroVir_CAF1 1644 110 - 1 UAUGAGCGCAUGCAGCAGCUGGAACAGAAGCUGGCCACCGUACUGCUGCAGGCUUUCGAGCAGUGCCACAGUUGGAUCCACCUGCUCAAGCUGACGAUUAUGUUUGGCUG ...(((.((.((((((((.(((..(((...)))....)))..)))))))).)).)))(((((((((((....)))...)).)))))).(((..((......).)..))). ( -36.20) >DroEre_CAF1 1378 110 - 1 UUUAGCCGAAUGCAACUGCUGGAGCAGAAGCUGGCCACGGUGUUGCUCCAGGCCUUUGAACAAUGUCACAGCUGGAUGCACUUGCUCCGACUGACUUUGAUGUUUGGCUC ...((((((((.(((..((((((((((..((((....)))).)))))))).))..)))..(((.((((..(.((((.((....)))))).))))).)))..)))))))). ( -40.00) >DroYak_CAF1 1388 110 - 1 UUUAGCCGAAUGCAACUGCUGGAGCAGAAGCUGGCCACCGUUUUGCUGCAGGCCUUUGAACAAUGUCACAGCUGGAUGCACUUGCUCCGGCUGACUUUGAUGUUCGGCAC ....(((((((((..((((....))))..)).((((...((......)).))))......(((.((((..((((((.((....)))))))))))).)))..))))))).. ( -42.70) >DroMoj_CAF1 1892 110 - 1 UUCCAGCGCAUGCAGCAACUGGAGCAGAAGCUGGCCACCGUGCUGCUGCAGGCCUUCGAGCAGUGCUACAGCUGGGUGCACCUGCUUAAGCUGACCCUGAUGUUCGGCUG ...(((((((.((((((..((((((....)))..)))...))))))))).......((((((......(((((....((....))...))))).......)))))))))) ( -41.02) >DroAna_CAF1 1200 110 - 1 UUCGGCAGGAUGCAGUUGCUGGAACAGAAACUGGCCACUGUCCUGCUUCAGGCUUUCGAGCAGUGUUACAGCUGGACGCACCUGCUCCGGCUGACACUGAUGUUUGGCGG ...((((((..(((((.((..(........)..)).)))))))))))....(((......((((((..((((((((.((....))))))))))))))))......))).. ( -48.20) >consensus UUCAGCCGAAUGCAGCUGCUGGAGCAGAAGCUGGCCACCGUGCUGCUGCAGGCCUUCGAACAAUGUCACAGCUGGAUGCACCUGCUCCGGCUGACUUUGAUGUUUGGCUC ..........((((((.(((((..(((...))).)))..))...)))))).(((...(((((......((((((((.((....)))))))))).......)))))))).. (-27.43 = -27.19 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:24 2006