
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,142,697 – 23,142,789 |
| Length | 92 |
| Max. P | 0.679108 |

| Location | 23,142,697 – 23,142,789 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -16.77 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
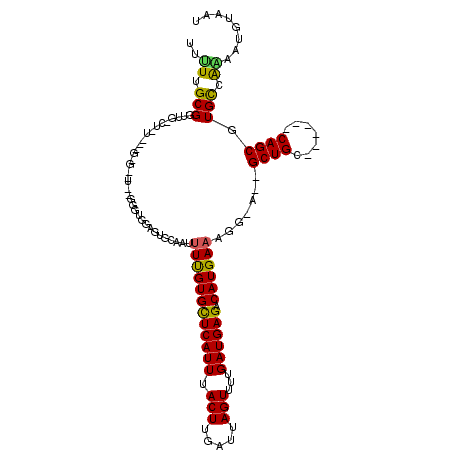
>3R_DroMel_CAF1 23142697 92 + 27905053 UUUUUGCGGUUG-CA-----------------GUUGUGCGAUUUCGUGCUCAUUUACUUGAUUAGUUCUGAUGAGACAUGAAAGG-A--GCUGC------CAGCGUGCCGAAAUGUAAU ......((((..-..-----------------((((.((..(((((((((((((.(((.....)))...)))))).)))))))..-.--))...------))))..))))......... ( -27.90) >DroVir_CAF1 23995 108 + 1 GCCUUGCGGUUA-CCGUG-CGCGAAUGCGGGUGAGCUAUAAAUUUGUGUUCAUUUACUUGAUUAGUUUUGAUGAGACAUGAAAGG-C--GCUGG------CAGCCUGCCAAAGUGUAAU (((((..(.(((-((.((-((....))))))))).).......(..((((((((.(((.....)))...))))).)))..)))))-)--..(((------(.....))))......... ( -30.10) >DroPse_CAF1 21085 115 + 1 UAUUUGCGGUUU-UUUUUUUGGGUUUUUGAGUGGAGUGCAAUUUUGUGCUCAUUUACUUGAUUAGUUUUGAUGAGACAUGAAAGG-A--GCUGCUGCUGGCAGCGUGUCGGAAAUUAAU .....(((((((-(((((.((..((.(..(((((((..((....))..)))...))))..)...........))..)).))))))-)--)))))..(((((.....)))))........ ( -29.00) >DroWil_CAF1 21423 92 + 1 UUUUUGCGGUUG-CU-----------------GU-GUGCGAUAUUGUGCUCAUUUACUUGAUUAGUUUUGAUGAGACAUGAAAGGUA--GCUGC------CAGCGUGUCAAAAUGCAAU .....(((((((-((-----------------(.-...)....(..((((((((.(((.....)))...)))))).))..)..))))--)))))------..((((......))))... ( -25.30) >DroMoj_CAF1 25669 111 + 1 GUCUUGCGGUUGGCUGUG-CUCAAAUGCGGGUGGGUUAUAAAUUCGUGUUCAUUUACUUGAUUAGUUCUGAUGAGACAUGAAAGG-CGCGCUGG------CAGCUUGCCAGUAUGUAAU ...(((((....((((.(-(.....(((.((((.(((.....((((((((((((.(((.....)))...))))).))))))).))-).)))).)------))....)))))).))))). ( -34.30) >DroPer_CAF1 20926 115 + 1 CAUUUGCGGUUU-UUUUUUUGGGUUUUUGGGUGGAGUGCAAUUUUGUGCUCAUUUACUUGAUUAGUUUUGAUGAGACAUGAAAGG-A--GCUGCUGCUGGCAGCGUGUCGAAAAUUAAU ............-........(((((((((...........(((..((((((((.(((.....)))...)))))).))..)))..-.--(((((.....)))))...)))))))))... ( -26.80) >consensus UUUUUGCGGUUG_CU_U___G_G__U__GGGUGGAGUGCAAUUUUGUGCUCAUUUACUUGAUUAGUUUUGAUGAGACAUGAAAGG_A__GCUGC______CAGCGUGCCAAAAUGUAAU ..((.(((..................................((((((((((((.(((.....)))...)))))).)))))).......((((.......)))).))).))........ (-16.77 = -15.88 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:21 2006