
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,141,673 – 23,141,799 |
| Length | 126 |
| Max. P | 0.999952 |

| Location | 23,141,673 – 23,141,775 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.06 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -9.07 |
| Energy contribution | -10.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
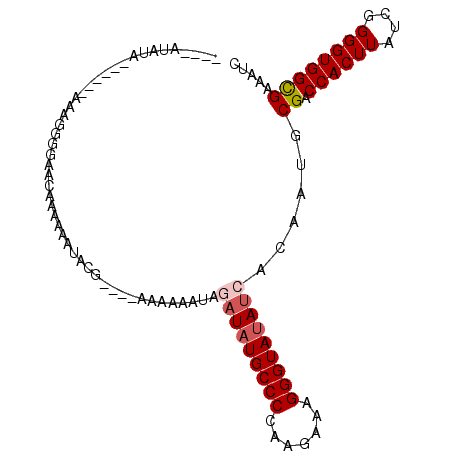
>3R_DroMel_CAF1 23141673 102 + 27905053 CAACUUAUGCACAUUAACUUAUCGGUAAUUUAUUGA-------AUAUA------AAAGGGGAACCAAAUAUACG----GAAGUAUAGAUAUGCCCUAAGAUAGGGUAAAUCGCAAUGCG .......(((...........((((((...))))))-------.....------...((....))...(((((.----...)))))(((.(((((((...))))))).))))))..... ( -24.70) >DroSec_CAF1 19299 102 + 1 UAACUCAUAAAAAUAAACUUAUCAAUAAUUUAUUGA-------AUAUA------AAAGGGGAACAAAAUAUACG----GAAGUAUAGAUAUGCCCUGAAAAAGGGUAUAUCGCAAUGCG ...(((.(((........)))((((((...))))))-------.....------...)))........(((((.----...)))))((((((((((.....))))))))))........ ( -24.30) >DroEre_CAF1 19064 99 + 1 CAGAUUUUAAAAA---GCUUAUGA-U-----ACGAA-------AUAUGACAAAAUCAGGGGGACACAAAAUACG----AAAAAAUAUAUAUGCCCCAAGAAAGGGUAUAUCACAAUGCG .............---((((.(((-.-----.((..-------...(((.....))).(....)........))----..........(((((((.......)))))))))).)).)). ( -13.20) >DroYak_CAF1 20956 110 + 1 UAGAUUUUAAAAA---GCUUAUAA-U-----AUGAAGUUUUGUAUUUGAGGUAAUCAGUAAUCAACAUAAAAGGGAACAAUAAAUAUAUAUGCCCCAAGAAAGGGUAUACCACAAUGCG ..........(((---((((....-.-----...)))))))(((((.(.((((...................(....)............(((((.......))))))))).)))))). ( -17.10) >consensus CAAAUUAUAAAAA___ACUUAUCA_U_____AUGAA_______AUAUA______AAAGGGGAACAAAAAAUACG____AAAAAAUAGAUAUGCCCCAAGAAAGGGUAUAUCACAAUGCG ......................................................................................(((((((((.......)))))))))........ ( -9.07 = -10.08 + 1.00)



| Location | 23,141,709 – 23,141,799 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.05 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -17.29 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.81 |
| SVM RNA-class probability | 0.999952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23141709 90 + 27905053 ----AUAUA------AAAGGGGAACCAAAUAUACG----GAAGUAUAGAUAUGCCCUAAGAUAGGGUAAAUCGCAAUGCGACCACUUAUUGGGGUGGCGAAAUC ----.....------...((....))...(((((.----...)))))(((.(((((((...))))))).)))....(((.(((.((....))))).)))..... ( -29.30) >DroSec_CAF1 19335 90 + 1 ----AUAUA------AAAGGGGAACAAAAUAUACG----GAAGUAUAGAUAUGCCCUGAAAAAGGGUAUAUCGCAAUGCGACCACUUAUCGGGGUGGCGAAAUC ----.....------..............(((((.----...)))))((((((((((.....))))))))))......((.((((((....))))))))..... ( -30.40) >DroEre_CAF1 19091 96 + 1 ----AUAUGACAAAAUCAGGGGGACACAAAAUACG----AAAAAAUAUAUAUGCCCCAAGAAAGGGUAUAUCACAAUGCGACCACUUAUCGGGGUGGUGAAUUC ----...............(....)........((----.........((((((((.......)))))))).......))(((((((....)))))))...... ( -18.59) >DroYak_CAF1 20986 104 + 1 UUGUAUUUGAGGUAAUCAGUAAUCAACAUAAAAGGGAACAAUAAAUAUAUAUGCCCCAAGAAAGGGUAUACCACAAUGCGCCCACUUAGCGGGGUGGCGAAAUA .((((((((.(((........))).........(....)..))))))))(((((((.......)))))))((((..(((.........)))..))))....... ( -21.00) >consensus ____AUAUA______AAAGGGGAACAAAAAAUACG____AAAAAAUAGAUAUGCCCCAAGAAAGGGUAUAUCACAAUGCGACCACUUAUCGGGGUGGCGAAAUC ...............................................(((((((((.......)))))))))......((.((((((....))))))))..... (-17.29 = -18.10 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:20 2006