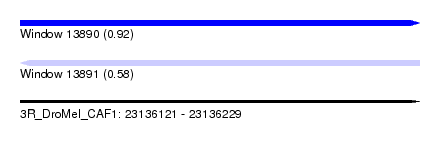
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,136,121 – 23,136,229 |
| Length | 108 |
| Max. P | 0.924727 |

| Location | 23,136,121 – 23,136,229 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -19.51 |
| Energy contribution | -21.73 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
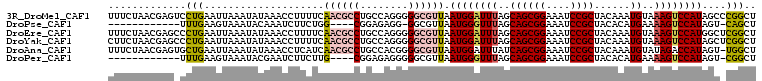
>3R_DroMel_CAF1 23136121 108 + 27905053 AGCCGGGCUAUGGACUUUACAUUUGUAGCGGAUUUCCGCUGCUAAAUCCAUUAACGCCCCCUGGCAGGCGUUGAAAAGGUUUAUAUUUAAUUCAGGACUCGUUAGAAA ...((((((.((((..........(((((((....)))))))((((((..((((((((........))))))))...)))))).......)))))).))))....... ( -31.50) >DroPse_CAF1 14517 90 + 1 AGCUG-ACUAUGGACUUUUCAUGUGUAGCGGAUUUCCGCUGCUAAACCCAUUAACGCC-CCUCUCCG----CCAGAAGAUUUGUAUUUACUUCAAA------------ .((..-...((((((.......))(((((((....))))))).....))))....)).-........----...((((...........))))...------------ ( -17.90) >DroEre_CAF1 13555 108 + 1 AGCCGAGCCAUGGACUUUACAUUUGUAGCGGAUUUCCGCUGCUAAAUCCAUUAACGCCCCCUGGCAGGCGUUGAAAAGGUUUAUAUUUAAUUCAGGGCUCGUUAGAAA ...((((((.((((..........(((((((....)))))))((((((..((((((((........))))))))...)))))).......)))).))))))....... ( -39.50) >DroYak_CAF1 15327 108 + 1 AGCCGAGCUAUGGACUUUACAUUUGUAGCGGAUUUCCGCUGCUAAAUCCAUUAACGCCCCCUGGCAGGCGUUGAAAAGGUUUAUAUUUAAUUCAGGGCUCGUUAGAAG ...((((((.((((..........(((((((....)))))))((((((..((((((((........))))))))...)))))).......)))).))))))....... ( -37.70) >DroAna_CAF1 13930 107 + 1 AGCCA-ACUAUGGUCUAUACAUUUGUAGCGGAUUUCCGCUGAUAAAUCCAUUAACGCCCCGUGGCAGGCGUUGAUGAGGUUUAUAUUUAAUUCAGCACUCGUUAGAAA ..(((-....)))((((.((.....((((((....))))))(((((((((((((((((........)))))))))).)))))))................)))))).. ( -33.40) >DroPer_CAF1 14395 91 + 1 AGCCG-ACUAUGGACUUUUCAUGUGUAGCGGAUUUCCGCUGCUAAACCCAUUAACGCCCCCUCUCCG----CAAGAAGAUUCGUAUUUACUUCAAA------------ .((..-...((((((.......))(((((((....))))))).....))))....))....(((.(.----...).))).................------------ ( -19.30) >consensus AGCCG_ACUAUGGACUUUACAUUUGUAGCGGAUUUCCGCUGCUAAAUCCAUUAACGCCCCCUGGCAGGCGUUGAAAAGGUUUAUAUUUAAUUCAGG_CUCGUUAGAAA ........((((((((((......(((((((....)))))))........((((((((........)))))))).))))))))))....................... (-19.51 = -21.73 + 2.23)



| Location | 23,136,121 – 23,136,229 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23136121 108 - 27905053 UUUCUAACGAGUCCUGAAUUAAAUAUAAACCUUUUCAACGCCUGCCAGGGGGCGUUAAUGGAUUUAGCAGCGGAAAUCCGCUACAAAUGUAAAGUCCAUAGCCCGGCU ..((....))((..((((...............))))..))..(((...((((....((((((((.(((((((....))))......))).)))))))).))))))). ( -28.36) >DroPse_CAF1 14517 90 - 1 ------------UUUGAAGUAAAUACAAAUCUUCUGG----CGGAGAGG-GGCGUUAAUGGGUUUAGCAGCGGAAAUCCGCUACACAUGAAAAGUCCAUAGU-CAGCU ------------.....................((((----(......(-((((((((.....)))))(((((....)))))...........))))...))-))).. ( -22.60) >DroEre_CAF1 13555 108 - 1 UUUCUAACGAGCCCUGAAUUAAAUAUAAACCUUUUCAACGCCUGCCAGGGGGCGUUAAUGGAUUUAGCAGCGGAAAUCCGCUACAAAUGUAAAGUCCAUGGCUCGGCU .......((((((.......................(((((((......))))))).((((((((.(((((((....))))......))).))))))))))))))... ( -37.20) >DroYak_CAF1 15327 108 - 1 CUUCUAACGAGCCCUGAAUUAAAUAUAAACCUUUUCAACGCCUGCCAGGGGGCGUUAAUGGAUUUAGCAGCGGAAAUCCGCUACAAAUGUAAAGUCCAUAGCUCGGCU .......(((((........................(((((((......))))))).((((((((.(((((((....))))......))).)))))))).)))))... ( -34.10) >DroAna_CAF1 13930 107 - 1 UUUCUAACGAGUGCUGAAUUAAAUAUAAACCUCAUCAACGCCUGCCACGGGGCGUUAAUGGAUUUAUCAGCGGAAAUCCGCUACAAAUGUAUAGACCAUAGU-UGGCU ..(((((((..((...........(((((.(.(((.(((((((......))))))).)))).))))).(((((....))))).))..))).))))(((....-))).. ( -26.70) >DroPer_CAF1 14395 91 - 1 ------------UUUGAAGUAAAUACGAAUCUUCUUG----CGGAGAGGGGGCGUUAAUGGGUUUAGCAGCGGAAAUCCGCUACACAUGAAAAGUCCAUAGU-CGGCU ------------...((((...........))))..(----(.((....(((((((((.....)))))(((((....)))))...........))))....)-).)). ( -23.60) >consensus UUUCUAACGAG_CCUGAAUUAAAUAUAAACCUUUUCAACGCCUGCCAGGGGGCGUUAAUGGAUUUAGCAGCGGAAAUCCGCUACAAAUGUAAAGUCCAUAGC_CGGCU .............(((....................((((((........)))))).((((((((..((((((....))))......))..))))))))....))).. (-15.97 = -16.75 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:18 2006