| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,128,234 – 23,128,347 |
| Length | 113 |
| Max. P | 0.936242 |

| Location | 23,128,234 – 23,128,347 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -41.18 |
| Consensus MFE | -24.67 |
| Energy contribution | -26.17 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
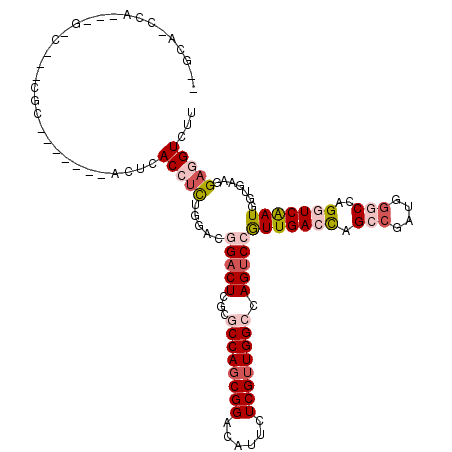
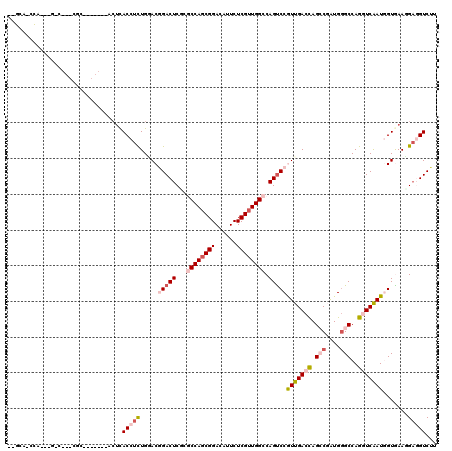
>3R_DroMel_CAF1 23128234 113 + 27905053 CCGCAACCACAUGGCCGAAGG-------ACUCACCUUUGGACGGACUCCUUCCAGCGGACAUUCUCGUUGGCCAGCCCAUUGACCAGCCGAUGGGCCAGGUCGAUGGUGAAGGCGGUCUU ((((...(((.((((((((((-------(.((.((...))...)).)))))).(((((......))))))))))...((((((((.(((....)))..)))))))))))...)))).... ( -47.50) >DroVir_CAF1 7061 100 + 1 --GCA--CU---------CUU-------ACUCACCUUUGCACCGACUCUCGCCAGCGGACAUUCUCGUUGGCCAGUCCGUUGACUAGCCUAUGCGCCAGAUCGAUGGUAAAGGACGUCUU --...--..---------...-------.....(((((((...((((...((((((((......)))))))).))))((((((((.((......)).)).)))))))))))))....... ( -32.50) >DroPse_CAF1 6464 117 + 1 UCGCACCCA---GACCGACGCACUCGACACUCACCUCUGGACGGACUCGCGCCAGCGGACAUUCUCGUUGGCCAGUCCGUUGACCAGACGAUGGGCCAGGUCAAUUGUGAAUGAGGUCUU ........(---((((....(((..(((.(((((.((((((((((((.(.((((((((......))))))))))))))))...))))).).))))....)))....))).....))))). ( -46.10) >DroMoj_CAF1 7513 105 + 1 --GUC--CACA-GGC---CAC-------GCUCACCUCUGCACAGACUCGCGCCAGCGGACAUUCUCGUUGGCCAGUCCAUUGACCAGCCUGUGGGCCAGAUCGAUGGUAAAGGACGUCUU --(((--((((-(((---...-------((........))...((((.(.((((((((......))))))))))))).........))))))))))......((((........)))).. ( -41.40) >DroAna_CAF1 5695 98 + 1 --GCACCUACA--------------------UACCUCUGAACGGACUCUUUCCACCGGAUGUUCUCGUUGGCCAGUCCGUUGACCAGCCGGUGGGCCAGGUCAAUGGUGAAUGAGGUCUU --..((((.((--------------------((((....((((((((....(((.(((......))).)))..))))))))((((.(((....)))..))))...)))..)))))))... ( -33.50) >DroPer_CAF1 6396 115 + 1 --GGAUACG---GACCGACGCACUCGACACUCACCUCUGGACGGACUCGCGCCAGCGGACAUUCUCGUUGGCCAGUCCGUUGACCAGACGAUGGGCCAGGUCAAUUGUGAAUGAGGUCUU --......(---((((....(((..(((.(((((.((((((((((((.(.((((((((......))))))))))))))))...))))).).))))....)))....))).....))))). ( -46.10) >consensus __GCA_CCA___G_C___CGC_______ACUCACCUCUGGACGGACUCGCGCCAGCGGACAUUCUCGUUGGCCAGUCCGUUGACCAGCCGAUGGGCCAGGUCAAUGGUGAAGGAGGUCUU ................................(((((.....(((((...((((((((......)))))))).)))))(((((((.(((....)))..))))))).......)))))... (-24.67 = -26.17 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:16 2006